Transcriptomic responses to environmental change in fishes: Insights from RNA sequencing
Abstract
Introduction
| Species | Tissue | Number of biological replicates | Experimental comparison | Environmental variable/challenge | Key processes involved in response | Reference |
|---|---|---|---|---|---|---|
| Temperature only | ||||||
| Acanthochromis polyacanthus | Liver | 4–5 | Developmental, transgenerational | Temperature (↑) | Metabolism (↑), immune response (↑↓), stress response (↑↓), tissue development (↑), transcriptional regulation (↑) | (Veilleux et al. 2015) |
| Cynoglossus semilaevis | Gill, liver, muscle | 3 | Long term | Temperature (↑) | Protein processing (↑), cell morphogenesis (↑), autophagy (↑), immune response (↓), hypoxic signalling (↑) | (Guo et al. 2016) |
| Hypomesus transpacificus | Whole larvae | 5 | Short term, interspecific | Temperature (↑) | Metabolism (↑), protein synthesis (↑), inducible transcription factors (↑), osmoregulation | (Jeffries et al. 2016) |
| Ictalurus sp. | Gill, liver | 1 (3) | Short term | Temperature (↑) | Oxygen transport (↑), protein folding and degradation (↑), metabolic process (↑), cytoskeletal organization (↑), protein synthesis (↓) | (Liu et al. 2013) |
| Melanotaenia duboulayi | Liver | 6 | Long term | Temperature (↑) | Immune response (↑), stress response (↑), developmental process (↑), metabolism (↓) | (Smith et al. 2013) |
| Oncorhynchus mykiss gairdneri | Gill | 3 (3) | Long term, intraspecific | Temperature (↑) | Stress response (↑), metabolism (↑), cellular process, response to stimuli | (Narum and Campbell 2015) |
| Pagothenia borchgrevinki | Liver | 3 | Short term | Temperature (↑) | Cell cycle (↓), ribosome biogenesis (↓), protein biosynthesis (↓) | (Bilyk and Cheng 2014) |
| Schizothorax richardsonii | Liver | 1 (3) | Short term | Temperature (↑) | Response to stimulus (↑↓), metabolic process (↑↓), protein folding and degradation (↑), immune response (↑), lipid metabolism (↑) | (Barat et al. 2016) |
| Spirinchus thaleichthys | Whole larvae | 5 | Short term, interspecific | Temperature (↑) | Stress response (↑), protein folding and degradation (↑), DNA damage (↑), aerobic metabolism (↑), osmoregulation | (Jeffries et al. 2016) |
| Squalius carolitertii | Muscle, liver, fin | 1 | Short term, interspecific | Temperature (↑) | Regulation of transcription (↑), RNA metabolism (↑), protein folding and degradation (↑), oxidation–reduction (↑↓) | (Jesus et al. 2016) |
| Squalius torgalensis | Muscle, liver, fin | 1 | Short term, interspecific | Temperature (↑) | Protein folding and degradation (↑), cell division (↓), DNA and RNA metabolism (↓), ribosome biogenesis (↓) | (Jesus et al. 2016) |
| Cyprinus carpio haematopterus | Brain, liver, spleen, gill, muscle | 1 (3) | Long term | Temperature (↓) | Protein localization and transport, cellular processes, signal transduction, genetic information processing, metabolism | (Liang et al. 2015) |
| Danio rerio | Whole larvae | 2 (50) | Short term | Temperature (↓) | Transcription (↑), metabolism (↑↓), transport (↑↓), phosphorylation (↑↓), cell motility (↓) | (Hung et al. 2016) |
| Whole larvae | 1 (50) | Developmental, short term | Temperature (↓) | RNA splicing and localization (↑), ribosome biogenesis (↑), protein catabolism (↑), metabolism (↓), oxidation–reduction (↓) | (Long et al. 2013) | |
| Brain, heart, liver, intestine, muscle, gill, spleen, kidney | 1 (20) | Short term | Temperature (↓) | Transcriptional regulation (↑), microtubule-based processes (↑), mRNA splicing (↑), proteolysis (↑), oxidation–reduction (↓) | (Hu et al. 2015a) | |
| Muscle | 4 | Developmental, long term | Temperature (↓) | Metabolism (↑), oxidation–reduction (↑), angiogenesis (↑), muscle contraction and remodelling (↑↓), translation (↓) | (Scott and Johnston 2012) | |
| Lates calcarifer | Muscle | 1 (8) | Long term, intraspecific | Temperature (↓Northern; ↑Southern) | Northern: microtubule-based process (↑), response to stress (↑); Southern: complement system (↓), cellular stress response (↑) | (Newton et al. 2013) |
| Neogobius melanostomus | Liver | 3 | Short term, interspecific | Temperature (↑↓) | Temperature (↑): cell cycle (↓), DNA replication (↓); temperature (↓): carboxylic acid metabolism (↑), amino acid transport (↑), protein catabolism (↑) | (Wellband and Heath 2017) |
| Proterorhinus semilunaris | Liver | 3 | Short term, interspecific | Temperature (↑↓) | Temperature (↑): immune response (↑); temperature (↓): detection of stimulus (↑), cell signalling (↑), regulation of gene expression (↑), immune response (↑) | (Wellband and Heath 2017) |
| Salinity only | ||||||
| Gymnocypris przewalskii | Gill, kidney | 6 | Intraspecific | Salinity | Response to stimulus, immune response, ion transport, cellular water absorption, neuroendocrine system | (Zhang et al. 2015) |
| Oryzias melastigma | Brain, liver, gonad | 1 (10) | Interspecific | Salinity | Ion transport, signalling, cell adhesion, metabolism | (Lai et al. 2015b) |
| Anguilla japonica | Corpuscle of Stannius gland | 2 | Long term | Salinity (↑) | Calcium metabolism (↑↓), blood pressure regulation (↑↓), ion transport (↑↓), cell adhesion (↑), morphogenesis (↑) | (Gu et al. 2015) |
| Gill | 2 | Long term | Salinity (↑) | Intracellular signalling cascade (↑), phosphate metabolic process (↑), regulation of cell proliferation (↑), cell adhesion (↑) | (Lai et al. 2015a) | |
| Esophagus | 1 | Long term | Salinity (↑) | Ion transport, cellular permeability | (Takei et al. 2017) | |
| Gasterosteus aculeatus | Kidney | 3 | Long term, intraspecific | Salinity (↑) | Ion transport, cellular water absorption | (Wang et al. 2014) |
| Gill | 10 | Short term | Salinity (↑) | ATP production (↑), signalling (↑), osmoregulation (↑), cellular permeability (↑) | (Kusakabe et al. 2017) | |
| Brain | 88–108 | Short term | Salinity (↑) | Hyperosmotic response, immune response | (Ishikawa et al. 2017) | |
| Lateolabrax maculatus | Liver | 3 | Long term | Salinity (↑) | Metabolites and ion transporters (↑), energy metabolism (↑), signal transduction (↑↓), immune response (↑↓), structure reorganization (↑) | (Zhang et al. 2017) |
| L. calcarifer | Intestine | 1 (3) | Short term | Salinity (↑) | Immune response (↑↓), signal transduction (↑↓), metabolism (↓), ribosome biosynthesis (↓) | (Xia et al. 2013) |
| Oreochromis mossambicus | Gill | 1 (4) | Long term | Salinity (↑) | Ion transport (↑), cell cycle (↑), metabolism (↑), signalling (↑↓), cellular remodelling (↓) | (Lam et al. 2014) |
| Oreochromis niloticus | Hepatopancreas | 1 (8) | Long term | Salinity (↑) | Amino acid, sterol, and protein metabolism (↑), immune response (↑↓), lipid metabolism (↓), signal transduction (↑) | (Xu et al. 2015) |
| Oryzias latipes | Intestine | 5 | Short term | Salinity (↑) | Protein phosphorylation, transcription regulation (↑), cell adhesion, signal transduction | (Wong et al. 2014) |
| Pangasianodon hypophthalmus | Gill, kidney, intestine | 1 (3) | Long term | Salinity (↑) | Apoptosis (↑), energy metabolism (↑), ion transport (↑↓), cellular reorganization (↑), signal transduction (↑↓) | (Nguyen et al. 2016) |
| Salvelinus alpinus | Gill | 6 | Long term | Salinity (↑) | Ion transport (↑↓), immune response (↑↓), cell cycle (↑), stress response (↑), developmental process (↓) | (Norman et al. 2014a) |
| Gill | 6 | Long term, intraspecific | Salinity (↑) | Immune response (↑↓), regulation of protein transport (↑) | (Norman et al. 2014b) | |
| Alosa pseudoharengus | Gill | 3 | Long term, intraspecific | Salinity (↑↓) | Landlocked/salinity (↓): freshwater ion uptake (↑), cellular permeability (↑); Anadromous/salinity (↑): ion secretion (↑) | (Velotta et al. 2017) |
| G. aculeatus | Gill | 5 | Long term, intraspecific | Salinity (↑↓) | Ion transport (↑↓), carbohydrate metabolism (↑↓), lipid metabolism (↑↓), rRNA processing (↓; salinity (↓) only), epithelial cell migration (↑↓) | (Gibbons et al. 2017) |
| Dissolved oxygen only | ||||||
| Ictalurus punctatus | Gill | N/A | Short term | Dissolved oxygen (↓) | Cellular permeability | (Sun et al. 2015) |
| Gill | N/A | Short term | Dissolved oxygen (↓) | Apoptosis (↓) | (Yuan et al. 2016) | |
| Larimichthys crocea | Brain | 6 | Short term | Dissolved oxygen (↓) | Neuroendocrine–immune system (↑↓), glycolysis (↑), protein synthesis (↓), aerobic metabolism (↓) | (Ao et al. 2015) |
| Megalobrama amblycephala | Liver, gill | 1 (3) | Long term | Dissolved oxygen (↓) | Hypoxic signalling (↑), angiogenesis, coagulation, DNA damage signalling and repair, metabolism | (Li et al. 2015b) |
| Morone sp. | Hepatopancreas | 3 (3) | Short term, long term | Dissolved oxygen (↓) | Lipid utilization (↑↓), metabolism (↑↓), autophagy (↑), apoptosis (↓) | (Beck et al. 2016) |
| O. melastigma | Brain, liver, gonad | 1 | Long term | Dissolved oxygen (↓) | Regulatory miRNAs of unknown biological function | (Lau et al. 2014) |
| Gonad | 2 (3) | Long term | Dissolved oxygen (↓) | Stress response, cell cycle, epigenetic modification, sugar metabolism, cell motility | (Tse et al. 2016) | |
| Brain | 2 (3) | Long term | Dissolved oxygen (↓) | Brain development (↑↓), nervous system development (↑↓), synaptic transmission (↑↓), axon guidance (↑↓), potassium ion transport (↑↓) | (Lai et al. 2016b) | |
| Gonad | 2 (3) | Long term | Dissolved oxygen (↓) | Steroidogenesis (↑) | (Lai et al. 2016a) | |
| pH only | ||||||
| Leuciscus waleckii | Gill, liver, kidney | 1 (9–10) | Intraspecific | pH (↑) | Metabolism (↑), immune response (↑), response to stimulus (↑↓), oxidation–reduction (↑↓), signalling | (Xu et al. 2013) |
| Sebastes caurinus | Muscle | 3–4 | Long term, interspecific | pH (↓) | Transcriptional regulation (↑), signalling (↑), stress response (↑) | (Hamilton et al. 2017) |
| Sebastes mystinus | Muscle | 2–3 | Long term, interspecific | pH (↓) | Muscle contraction (↑↓), signalling (↑↓), metabolism (↑↓), cellular structure (↑↓), transcription (↑↓) | (Hamilton et al. 2017) |
| Multiple stressors | ||||||
| O. mossambicus | Spleen | 1 (6) | Long term | Temperature (↑)* | Oxygen metabolism (↑↓), energy metabolism (↑↓), hypoxic signalling (↑), immune response (↑↓) | (Wang et al. 2016) |
| Chanos chanos | Brain, gill, liver, kidney | 1 (8) | Developmentala, long termb | Salinitya, temperatureb (↓) | Metabolism (↑↓) | (Hu et al. 2015b) |
| D. rerio | Whole larvae | 1 (50) | Developmentalc, short termd | Temperaturecd (↓), dissolved oxygencd (↓) | Oxidation–reduction (↑), oxygen transport (↑), hemoglobin biosynthesis (↑), ion transport (↑), fatty acid biosynthesis (↑) | (Long et al. 2015) |
| Colossoma macropomum | Muscle | 1 (6) | Short term, long term | Temperature (↑), pH (↓) | Metabolism (↑↓), development (↑), cellular organization (↑), macromolecule biosynthesis (↑↓), translation (↓) | (Prado-Lima and Val 2016) |
| P. borchgrevinki | Gill | 5 | Long term | Temperature (↑), pH(↓) | Immune response (↑), stress response (↑↓), cell proliferation (↓), cell death (↑), protein folding and degradation (↑) | (Huth and Place 2016a) |
| Trematomus bernacchii | Gill | 5 | Long term | Temperature (↑), pH (↓) | Immune response (↑), cell death (↑), carbohydrate and lipid metabolism (↑↓), signal transduction (↓), cell proliferation (↓) | (Huth and Place 2016b) |
| Alcolapia grahami | Gill | 5 | Long term, interspecific | pH (↑), salinity (↑), temperature (↑), dissolved oxygen (↑↓) | Energy metabolism (↑), ion transport (↑), stress response (↑), immune response (↑), osmoregulation (↑↓) | (Kavembe et al. 2015) |
Note: The number of biological replicates is given with the number of individuals pooled within each replicate denoted in parentheses when applicable. Key processes involved in responses include those of focal interest to the authors, those to which the greatest number of dysregulated genes were annotated, and those that showed the highest enrichment, to a maximum of five processes. Arrows indicate the relationship between environmental and response variables, where applicable. Identical superscripts between the “Experimental comparison” and “Environmental variable/challenge” columns denote which comparisons were made with which variables when multiple options are present within a study. Asterisks denote that the challenge was conducted in combination with a bacterial infection.
Plastic responses to environmental change
Short-term, acute responses
Long-term, chronic responses
Developmental plasticity
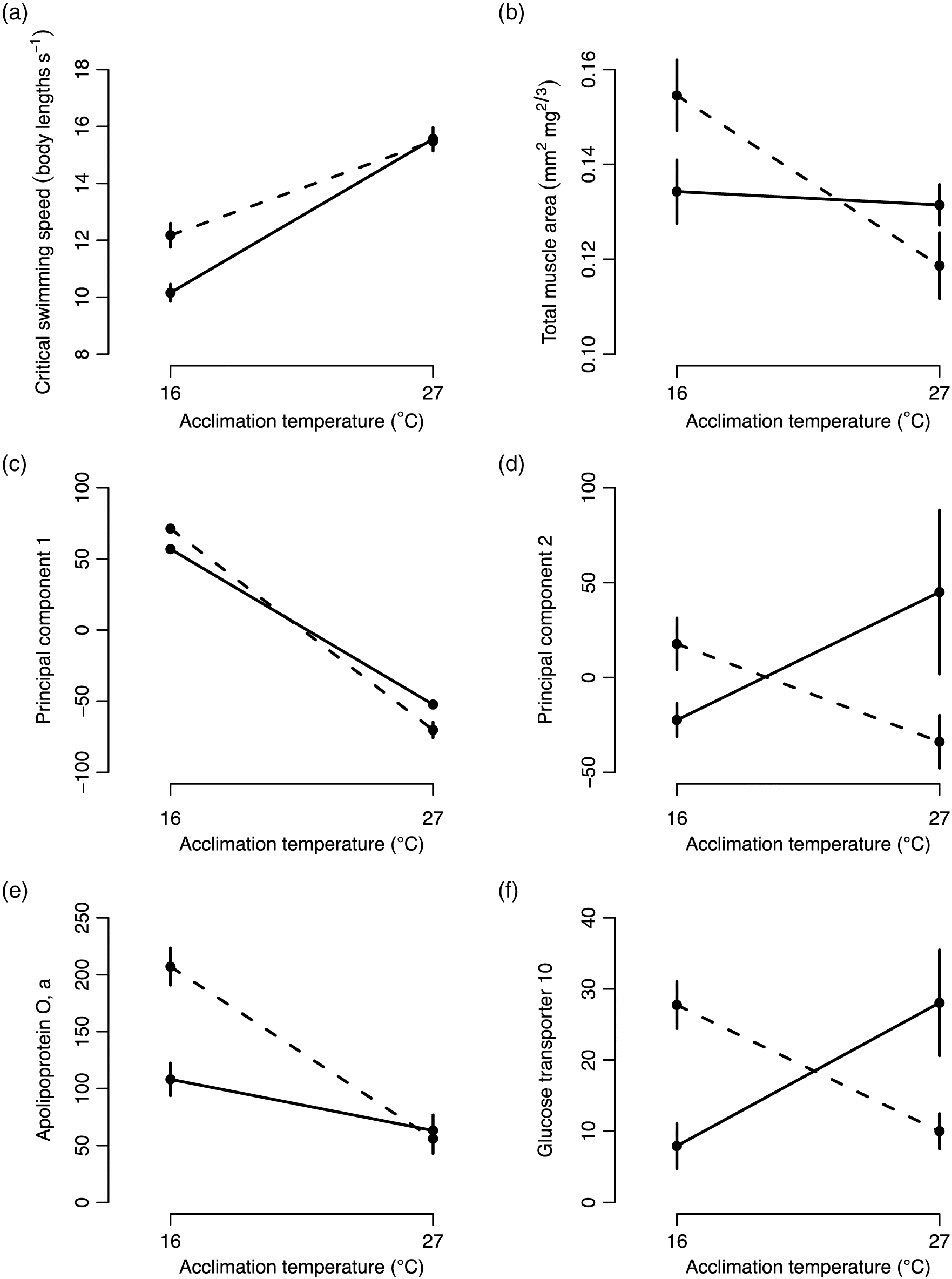
Transgenerational plasticity

Responses to multiple stressors
Evolutionary responses to environmental change
Identifying candidate genes for adaptation
Intraspecific variation in transcriptomes
Population-level variation in transcriptional plasticity

Family-level variation in transcriptional plasticity
Challenges and directions for future research
Experimental protocols and sampling design
Bioinformatic analysis
Conceptual challenges
Conclusion
Acknowledgements
References
Information & Authors
Information
Published In

History
Copyright
Data Availability Statement
Key Words
Sections
Subjects
Plain Language Summary
Authors
Author Contributions
Competing Interests
Metrics & Citations
Metrics
Other Metrics
Citations
Cite As
Export Citations
If you have the appropriate software installed, you can download article citation data to the citation manager of your choice. Simply select your manager software from the list below and click Download.
