Competition during enrichment of pathogenic Escherichia coli may result in culture bias
Abstract
Deadly outbreaks and illnesses due to Shiga toxin-producing Escherichia coli (STEC) occur worldwide; however, the cultivation methods required for adequate monitoring and traceback investigations are inefficient at best. Detection of STEC relies heavily on enrichment; yet no standard media or protocols exist. Furthermore, whether enrichment may bias detection of multiple STEC serogroups from complex samples is unknown. Thus, 14 STEC strains of serogroups O157 and the top six non-O157s (O26, O45, O103, O111, O121, and O145) were enriched in pairs for 6–78 h in broth and evaluated by quantitative polymerase chain reaction (qPCR). Here we show that a conventional 6-h enrichment protocol did not result in intra-species culture bias for the isolates tested. However, subsequent enrichments often produced biased cultures, with differences in the qPCR gene copy number ≥2 log10 apparent in 12%, 38%, and 52% of competitions after 30, 54, and 78 h of consecutive enrichments, respectively. Some strains were able to prevail and (or) out-compete the opponent strain in 100% of competitions. Our results suggest that culture bias should be considered and (or) evaluated further due to the potential implications during routine pathogen screening and outbreak investigations.
Introduction
Infection with Shiga toxin-producing Escherichia coli (STEC) ranges from mild to severe and can include diarrhea, hemorrhagic colitis, hemolytic uremic syndrome, and death. Although E. coli O157:H7 has been subjected to international reporting for nearly two decades, the most prevalent non-O157 serogroups have only recently gained attention (Gill and Gill 2010). Over 435 STEC serotypes have been isolated from cattle (the main reservoir of these pathogens), of which 12–17% are associated with human illness (Mathusa et al. 2010; Kaspar et al. 2010).
STEC are often present at low ratios within the total bacterial population. To exceed the threshold of detection, isolation requires enrichment and selective media. Enrichment can increase cell numbers to achieve a detectable level in both culture and molecular-based assays (at least 100 cells/g food or feces) (Possé et al. 2008; Bai et al. 2012; Paddock et al. 2012). Standard detection procedures and selective media have been developed for O157-STEC; however, no gold standard for the cultivation and isolation of non-O157 STEC exists. Isolation of non-O157 STEC from complex matrices such as food or feces is often impaired due to high levels of background microbiota, growth inhibitors, and a lack of selective media. Additionally, samples hosting multiple pathogenic strains complicate detection measures and relevant strains may not be identified (Gorski 2012; Bettelheim 2007).
Several enrichment protocols for STEC have been reported, each utilizing different combinations of media, supplements, incubation temperatures, and assessment times. Yet no medium or protocol has proven appropriate for all serogroups or sample types (e.g., food, feces, and water) (Durso 2013). Among current protocols, typical incubation temperatures range from 35 to 42 °C, and duration from 6 to 24 h (Vimont et al. 2006; Sidari and Caridi 2011). Although higher enrichment temperatures may be more selective for some STEC strains, longer incubations are sometimes applied simply for practical reasons in the laboratory such as allowing incubations to run overnight to avoid removing cultures from the incubator during non-typical work hours (Vimont et al. 2006). Any enrichment should provide a balance between recovery of the target organism while avoiding the overgrowth of competing cells (Gorski 2012). Consequently, enrichment broths are frequently modified by the addition of antibiotics to suppress the growth of background microflora. However, O157 and non-O157 STEC are a diverse group, and the addition of antibiotics has been shown to inhibit the growth of some STEC strains (Sidari and Caridi 2011; Vimont et al. 2007).
Culture bias has been described between strains of the same species or subgroup for Listeria monocytogenes and Salmonella spp. (Gorski 2012). Studies using Salmonella strains have reported dissimilar recovery characteristics in enrichment media leading to isolation of certain strains over others (Gorski 2012; Singer et al. 2009). Similarly, variants naturally present in populations of E. coli O157:H7 show differences in stress resistance that affected phenotypes isolated after 6-h enrichment (Carter et al. 2011). Another study showed that numerous STEC serogroups are concurrently present within pooled cattle feces and that prolonged storage time prior to enrichment of feces significantly decreased the number of serogroups detected, indicating that the population of E. coli in cattle feces is constantly changing even without enrichment (Conrad et al. 2014). In the same study, numbers of STEC O26 inoculated into fresh feces did not increase over the course of enrichment (as quantified by PCR), suggesting that this strain lacked the ability to effectively compete with existing microbiota in fecal samples during enrichment (Conrad et al. 2014). Still, it is unknown whether a culture bias arises during enrichment of multiple STEC serogroups, which could result in certain strains being more (or less) likely to be isolated than others. The principles governing competitive growth of these bacteria are not well understood, and in vitro experiments are scarce. The objectives of this study were to determine: if serogroup- or strain-specific competition (culture bias) exists between STEC during controlled enrichment of mixed cultures; whether this competition changed over time in the course of six subsequent enrichments; and lastly, whether toxin and virulence gene profiles influenced a STEC strain’s ability to compete during enrichment.
Methods
Bacterial cultures, media, and culture conditions
All STEC isolates used in this study were obtained from cattle feces at shipment to slaughter (Conrad et al. 2014). Isolates were selected for this study based on the virulence gene profile (Table 1). Group 1 isolates possessed both stx1 and stx2 (aside from serogroup isolate O26), whereas group 2 isolates had only one stx gene (stx1 or stx2). Group 1 isolates are designated by serogroup followed by -1 (e.g., O26-1), whereas isolates from group 2 are designated by serogroup followed by -2. Non-O157 STEC were streaked from glycerol stocks onto MacConkey agar (MAC), and O157 isolates were streaked onto sorbitol MacConkey agar containing cefixime and tellurite (CT-SMAC; Dalynn Biologicals, Calgary, Canada). Plates were incubated overnight (~16 h) at 37 °C. A single colony was selected from each overnight plate and inoculated into 10 mL E. coli (EC) broth (EMD Millipore, Etobicoke, Canada). Overnight broth cultures were incubated statically at 37 °C. A flowchart representation of the methodology is given in Fig. 1.
Table 1.
| Serogroup | Group | stx1 a | stx2 b | eae c | ehxA d | repA e |
|---|---|---|---|---|---|---|
| O26 | 1 | ✓ | ✓ | ✓ | ✓ | |
| O26 | 2 | ✓ | ✓ | ✓ | ||
| O45 | 1 | ✓ | ✓ | ✓ | ✓ | |
| O45 | 2 | ✓ | ✓ | ✓ | ✓ | |
| O103 | 1 | ✓ | ✓ | ✓ | ✓ | |
| O103 | 2 | ✓ | ✓ | ✓ | ✓ | |
| O111 | 1 | ✓ | ✓ | ✓ | ✓ | ✓ |
| O111 | 2 | ✓ | ✓ | ✓ | ✓ | |
| O121 | 1 | ✓ | ✓ | ✓ | ✓ | ✓ |
| O121 | 2 | ✓ | ✓ | ✓ | ✓ | |
| O145 | 1 | ✓ | ✓ | ✓ | ✓ | ✓ |
| O145 | 2 | ✓ | ✓ | ✓ | ✓ | |
| O157 | 1 | ✓ | ✓ | ✓ | ✓ | ✓ |
| O157 | 2 | ✓ | ✓ | ✓ | ✓ |
a
stx1—Shiga toxin 1 gene.
b
stx2—Shiga toxin 2 gene.
c
eae—intimin gene.
d
ehxA—EHEC enterohemolysin gene A.
e
repA—replication initiation gene A.
Fig. 1.
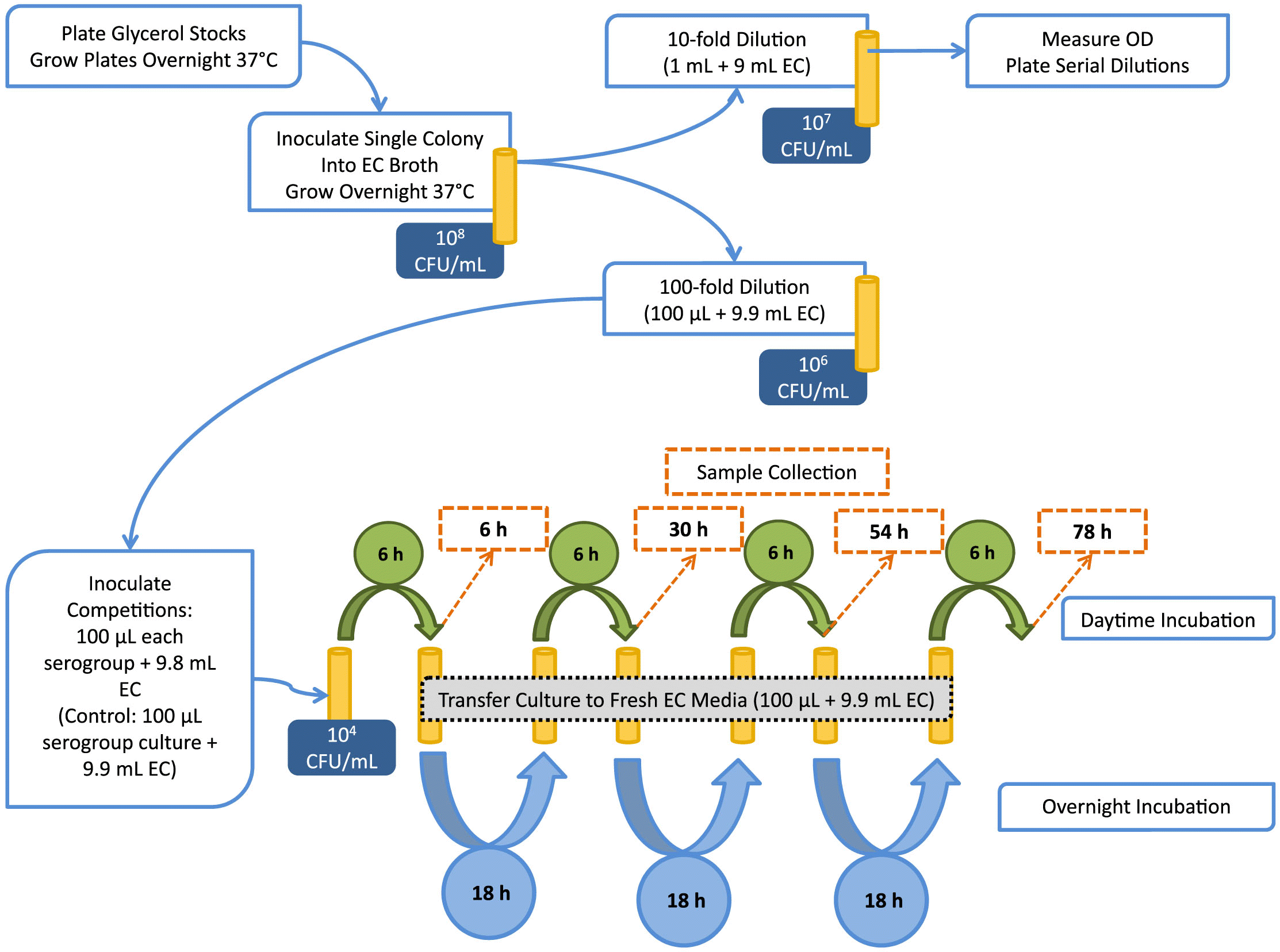
Dual-serogroup competitions and preparation of enrichment cultures
In a Latin square design, each of the group 1 and group 2 isolates (Table 1) was matched against each of the other isolates within that group in two-serogroup competitions (trial 1 and trial 2, respectively). Finally, select isolates from group 1 and group 2 were assessed in trial 3. Competitions were completed in both EC broth and diluted (50%) EC broth (1:1 broth: water).
Each overnight culture was diluted 10-fold in EC, and optical density (OD600) was measured photometrically using a Genesys-20 spectrophotometer (Thermo Scientific, Waltham, USA). Additional 10-fold serial dilutions were prepared in phosphate-buffered saline, and 100 µL of 10−5 and 10−6 dilutions were plated on CT-SMAC or MAC and incubated overnight at 37 °C.
For dual-serogroup inoculations (Fig. 1), each overnight culture was diluted 100-fold in EC (except for isolate O121-1, which had a lower OD and was adjusted by 10-fold dilution prior to inoculation). Then, 100 µL of each diluted culture was added to 9.8-mL EC or 50% EC broth. For controls, 100 µL of diluted pure culture was added to 9.9-mL broth. Each culture (including dual-serogroup enrichments and pure-culture controls) contained 104 CFU/mL of each strain at time 0.
Cultures were incubated statically for 6 h at 37 °C, at which time 1 mL of culture was removed and centrifuged at 11 000g for 10 min. Supernatant was removed and cell pellets were stored at –20 °C until DNA isolation using the NucleoSpin Tissue Kit (Macherey-Nagel, Bethlehem, USA) for trial 1. For subsequent trials, 200 µL of culture was removed, pelleted as above, and supernatant removed. These cells were re-suspended in 50 µL 1 × TE (10 mM Tris, pH 8.0) and heat-lysed at 95 °C for 5 min in order to minimize costs related to extraction.
Another 100 µL of the mixed 6 h culture was transferred to 9.9-mL fresh broth and incubated statically over night at 37 °C (Fig. 1). This process was repeated over four consecutive days and three nights. The intermediate samples were taken after 30 and 54 h of total enrichment. The final sample was taken 78 h after initial inoculation, and after seven alternating incubations of 6 and ~18 h.
Detection of serogroups by quantitative PCR
Escherichia coli were enumerated by quantitative polymerase chain reaction (qPCR) detection of serogroup-specific wzx/wzy gene fragments (primer and probe sequences given in Table 2). Reaction mixtures contained 1× QuantiFast Multiplex PCR NoROX Master Mix (Qiagen, Toronto, Canada), 0.5 µM of each primer, 0.2 µM of the probe, 2 µL of the template DNA, and nuclease-free water to a total final volume of 25 µL. Each qPCR was compared with standards containing 10–108 copies (described previously by Conrad et al. (2012)) and positive and negative controls in duplicate. Thermocycling conditions included an initial activation step at 95 °C for 5 min, followed by 45 cycles at 95 °C for 30 s and at 60 °C for 30 s. All qPCR reactions were completed in duplicate, and the gene copy numbers are reported as averages. In addition, isolates were screened by PCR for virulence genes, including Shiga toxins and pathogenicity island/locus of enterocyte effacement (LEE)-related genes (Conrad et al. 2014).
Table 2.
| Serogroup | Namea | Sequence (5′–3′) | Amplicon size (bp) |
|---|---|---|---|
| O26 | O26-F-50b | ATTGCAGCGCCTATTTCAGC | 200 |
| O26-R-50b | ATTAGAAGCGCGTTCATCCCT | ||
| O26-P-52 | GTTGAAACACCCGTAATGGCCACT [TQ3~TF3] | ||
| O45 | O45-F-50b | GATCTGTGGAGCCGAGATGG | 250 |
| O45-R-50b | TTTGAGACGAGCCTGGCTTT | ||
| O45-P-52 | TGCTGCAAGTGGGCTGTCCA [TQ2~TF2] | ||
| O103 | O103-F-50b | ATCTTCTTGCGGCTGCAGTT | 190 |
| O103-R-52 | ATGGGGGTGATTGGAGCGTT | ||
| O103-P-52 | ACAGCTTGCCAATATCCGGCATCC [TQ3~TF3] | ||
| O111 | O111-F-50b | CGCAAGACAAGGCAAAACAGA | 243 |
| O111-R-52 | ACTGGTGGCGTCTCACTTAGTT | ||
| O111-P-52 | AGGCAAGGGACATAAGAAGCCAGT [TQ2~TF2] | ||
| O121 | O121-F-52 | GGCATTCCTCAGTATCTTCTTGTTAAG | 250 |
| O121-R-50b | AGCAAGCCAAAACACTCAACA | ||
| O121-Pc | TTAACACGGGCGTGGTTGGA [TQ3~TF3] | ||
| O145 | O145-F-51b | GCGGGTGTTGCCCGTTCTGT | 216 |
| O145-R-52 | ACCTGGCATGCTCCCCTCCT | ||
| O145-P-52 | ACCGCCTGGAGTTGGGGCTT [TQ2~TF2] | ||
| O157 | O157-F2b | AGGGGTTGTATGCTCGTTGT | 121 |
| O157-R2b | TGGAACACCTTCAACTTGCTCT | ||
| O157-Pb | GGACAAGACGGAGAACAAAATGACTCA [TQ3~TF3] |
a
Primers and probes were designed in this study except where otherwise indicated.
Statistical analyses
Copy numbers were log transformed prior to analyses using the mixed model procedure of SAS, with serogroup, media (EC or diluted EC), repA and stx gene profiles of each isolate as independent variables and isolate as a repeated variable. “Winners” (colonies with enhanced fitness (FE)) were defined as the isolate with the higher serogroup-specific copy number by qPCR. Subsequently, records of FE (wins) and records of depressed units (losses) for each isolate were analyzed using the Glimmix procedure (SAS) in a binary distribution with replicate as a random variable and isolate, serogroup, media, repA and stx gene profiles as independent variables.
Results
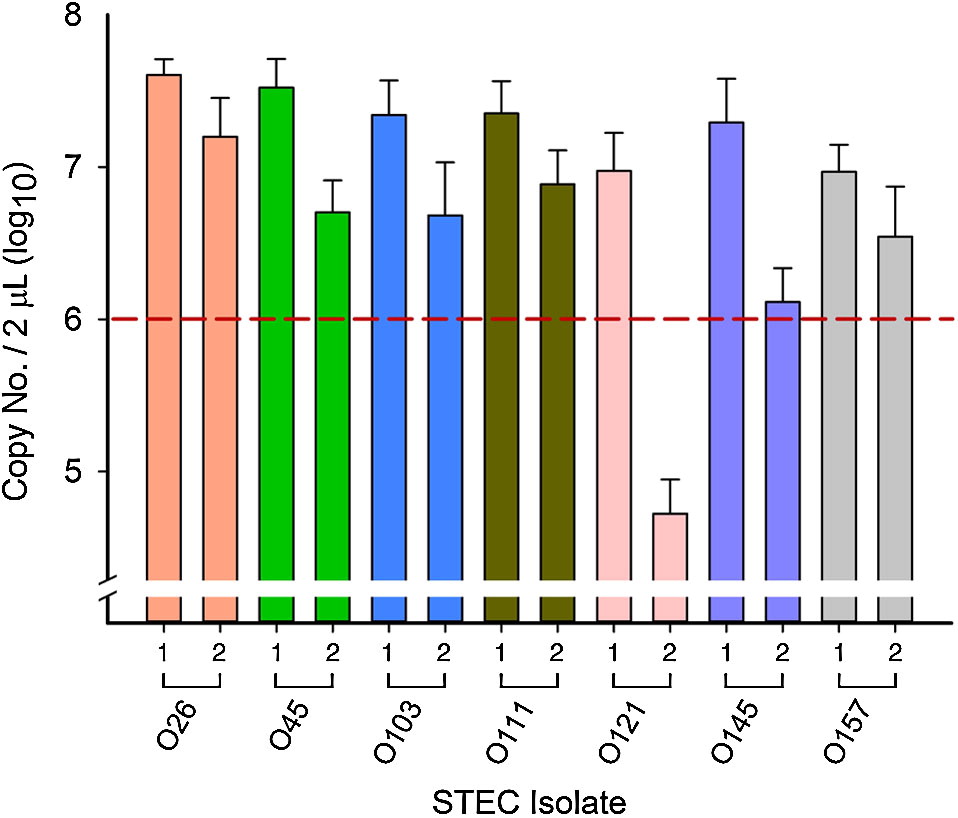
All overnight cultures used in inoculations reached 108 CFU/mL and optical densities for 10-fold dilutions of overnight pure cultures ranged from 0.104 to 0.158 (107 CFU/mL), with the exception of O121-1 (0.028–0.033; 106 CFU/mL). When the glycerol stock of O121-1 was first plated, colonies appeared beige (indicating nonlactose fermenting). Following the first overnight incubation in EC broth and re-plating of serial dilutions, approximately 15% of O121-1 colonies appeared pink indicating a shift toward lactose fermentation. PCR was used to confirm that both pink and beige colonies were O121-1 and not the result of contamination.
After the first 6 h of incubation, all isolates reached 106–107 copies per 2 µL extracted DNA (relation of copy number/mL to CFU/mL has been described previously by Conrad et al. (2014)) across all competitions, with the exception of O121-2 (Fig. 2). Although the initial OD reading was similar to all other isolates, O121-2 reached 104 copies per 2 µL extracted DNA in all competitions (Fig. 2), a difference of 2 log10 or more compared with other isolates from all other serogroups.
Fig. 2.
There were 57 individual competitions in each media (114 in total; Fig. 3). Differences in copy number ≥2 log10 were apparent in 12%, 38%, and 52% of competitions after 30, 54, and 78 h of consecutive enrichments, respectively (C. Conrad, personal observation, 2015). For remaining enrichments, copy numbers of the two strains were similar with ≤2 log10 difference between 30 and 78 h of incubation.
Fig. 3.
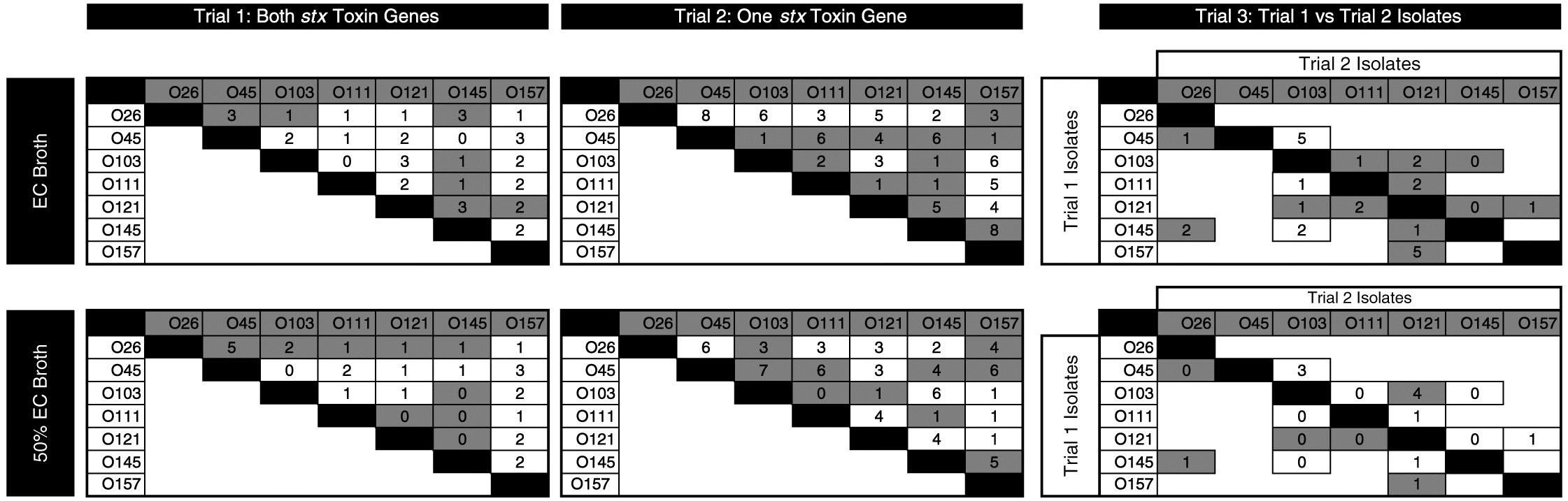
The isolates with FE from each competition for the three trials, after 78 h of sequential enrichments, are given in Fig. 3. The serogroup-specific copy numbers between competing isolates differed from less than 1 log10 to 8 log10 copies per 2 µL DNA.
For the majority of competitions, the FE strain was the same in both concentrated and diluted media. In cases where the dominant serogroup differed between media, one of the serogroups was often O121. The isolates with the highest number of competitions prevailed (as percentage) were O45-1, O26-2, and O145-1 (Fig. 4), with O45-1 and O26-2 prevailed more competitions than O26-1, O121-1, O45-2, and O157-1 (P < 0.05). Serogroup per se did not influence the outcome of competitions. Rather, the number of competitions prevailed was impacted by the individual isolate (P < 0.01). In other words, two isolates may share the same serogroup yet one isolate may be a strong competitor, and the other may be weak.
Fig. 4.
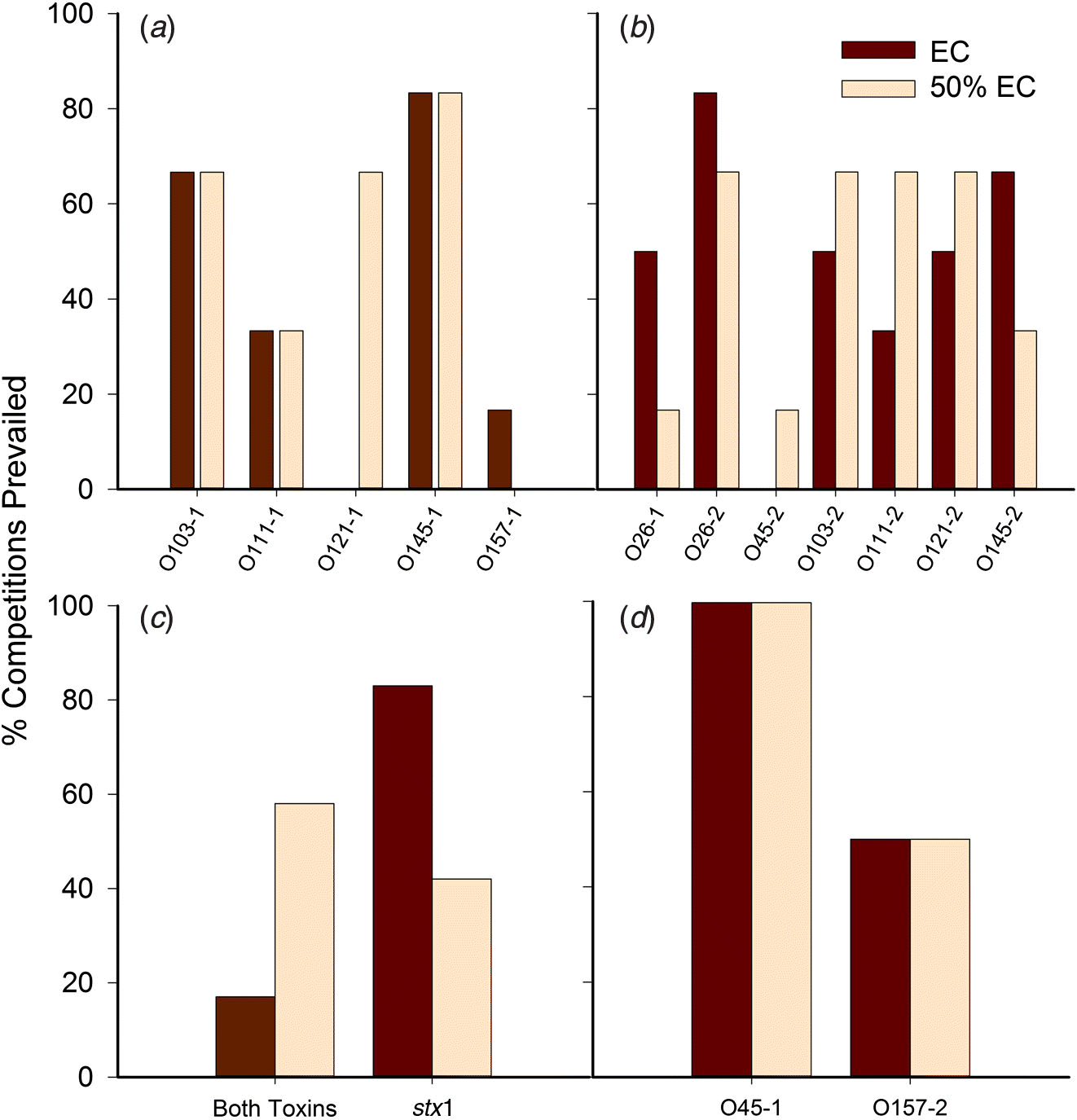
For trial 1, each isolate prevailed in a similar percentage of competitions irrespective of media, with the exceptions of O121-1, which prevailed in 60% more competitions in diluted EC broth (Fig. 4a), and O157-1, which was least able to compete, prevailing in less than 20% of competitions. For trial 2, isolate O26-2 prevailed in the greatest number of competitions, and O45-2 prevailed in the least (Fig. 4b).
The Shiga toxin profile of isolates had a significant impact on the outcome of the competition monitored by copy number (P < 0.01), whereas serogroup, media, and other virulence genes had no significant impact in all trials. If both isolates shared similar stx gene profiles (e.g., stx1 vs. stx1, stx2 vs. stx2, both toxin genes vs. both toxin genes), copy numbers for each were superior to those of stx1 matched against stx2-only isolates (P < 0.001), but did not significantly differ from copy numbers when other combinations of stx genes were present (e.g., stx1 vs. both, stx2 vs. both). A tendency for interaction between the media and the stx profile was demonstrated, as stx1-only isolates prevailed in 80% of competitions against those with both toxin genes (trial 3) in EC broth whereas isolates with both Shiga toxin genes prevailed in 60% of competitions against stx1 isolates in the diluted broth (Fig. 4c). Two isolates contained stx2 only: O45-1 and O157-2. The O45-1 isolate prevailed 100% of competitions in both media when competing against isolates with both Shiga toxin genes. However, when O157-2 competed against isolates with stx1 only, it prevailed 50% of competitions in both media (Fig. 4d).
Discussion
Studies have shown that enrichment can severely distort the apparent diversity or taxonomic profile within populations due to culturing biases (Dunbar et al. 1997; Pettengill et al. 2012). In one study, three agar media (Rainbow, Chromagar, and Blood agar) were compared for recovery of STEC from over 4000 enrichments (tryptic soy broth) of a variety of sample types (feces, plant, soil, and water). Out of 880 positive samples, no more than 56 were positive on all three media (Cooley et al. 2013). Furthermore, 377, 112, and 110 samples were positive on only one of the three media, suggesting a potential culture bias (Cooley et al. 2013).
Interestingly, reports of STEC prevalence in cattle feces show substantial variation, ranging from 0.2% to 70% (Hussein 2007; Kaspar et al. 2010). Such variance could be due to differences in climate, ecology, farming practice, and variation in sampling (Hussein 2007). However, such differences could also, in part, be a reflection of culture biases and differences in methodologies used for pathogen isolation. Enrichment of complex samples (food, feces, soil, etc.) could result in a biased culture due to the nature of the microbiota present, production of bacteriocins, bacteriophage interactions, differences in relative growth rates, and presence of growth inhibitors (Martínez-Castillo and Muniesa 2014; Muniesa et al. 2005; Pettengill et al. 2012). The aim of the present study was to utilize a simplified scenario to examine intra-species competition between STEC following enrichment using solely two bacterial strains in a broth medium. Competitive trends were examined between STEC strains based on molecular serogroup characteristics specific to O157 and the top six non-O157.
Numerous observations reported that E. coli cells in stationary phase can contain multiple chromosomes (Boye and Lobner-Olesen 1991; Akerlund et al. 1995). Despite this, qPCR was chosen to measure approximate cell number within mixed cultures due to speed and sensitivity of the technique. In comparison to PCR, traditional culturing methods are not specific enough to determine qualitatively and (or) quantitatively individual serogroups in mixed cultures. In the light of increasing chromosomes per cell, higher copy numbers are indicative of high cell concentrations, representing an adequate way to differentiate cell number among the used strains.
During the initial 6 h of enrichment, no significant differences were observed among the cell numbers of the competing strains within the same medium (diluted or undiluted). However, many of the subsequent enrichments (30, 54, and 78 h) produced biased cultures. Although enrichments longer than 30 h are not typical, the intent was to demonstrate the effect of prolonged storage of a sample prior to enrichment and analysis. Often, samples are collected days before subsequent analysis in the laboratory. In general, samples are shipped and held at 4 °C, but sometimes they are shipped under suboptimal storage conditions. Our results suggest that competition could be ongoing from the moment of sample collection, and delaying analysis could mean the results obtained are not reflective of the original sample.
The results from our test group of 14 strains do not suggest that any of the “Top 7” serogroups dominate all other serogroups during enrichment. Conversely, our results indicate that intra-species STEC competition is strain-specific. A study using Salmonella found similar results, which indicated that different selective media gave dissimilar patterns of strain dominance in both pure culture and fecal enrichment cultures (Gorski 2012). No serotype was most fit, but strains of certain serogroups were more likely to dominate than others.
The stx gene profile may in some way influence the ability of STEC strains to proliferate and coexist in the tested medium. Overall, strains possessing one stx toxin gene prevailed a greater number of competitions compared with those harboring both toxin genes. The majority of single toxin isolates contained stx1 only; however, two isolates possessed solely stx2, with one of these isolates prevailing 100% of its competitions. In undiluted broth, isolates with solely stx1 prevailed the greater number of competitions compared with isolates with both stx genes. However, when the nutrients within the media were restricted by 50%, isolates with both toxin genes prevailed nearly 20% more competitions than stx1-only isolates. This suggests that STEC strains may differ in their competitive ability when nutrient concentrations are limited, although statistical analyses did not show a significant influence of media, likely due to lack of replication.
The mechanisms and evolutionary forces governing the emergence of new STEC types are not well understood. Phylogenetic analyses have revealed that the gain and loss of virulence elements has occurred several times and in parallel in separate lineages (Franz et al. 2014). Such convergent evolution suggests the accumulation of specific combinations of virulence factors offers a selective advantage to the bacterium (Franz et al. 2014). Competitive advantages of E. coli possessing Shiga toxin genes have been reported. As one study demonstrates, Shiga toxins may act as a bacterial defense against Eukaryotic predators (Lainhart et al. 2009). Shiga toxins have also been implicated in the promotion of adherence and colonization of O157:H7 to human intestinal epithelial cells in comparison to stx-negative strains (Liu et al. 2010). However, selective advantages are more likely to operate in the bovine reservoir or external environment than in the human gastrointestinal tract as they are considered “accidental hosts,” and STEC/enterohemorrhagic E. coli (EHEC) are unlikely to have evolved specifically to cause human infection (Franz et al. 2014).
Laboratory-cultured bacteria are assumed to behave like those in nature. However, bacterial strains have been reported to rapidly transform during laboratory domestication as demonstrated by the O121 isolate in the present study, which alternated in ability to ferment lactose. Therefore, to decrease the likelihood of in-house domestication, potentially producing biased cultures, shorter and fewer incubations are recommended (Eydallin et al. 2014). Enrichment protocols other than the one in this study (different media, duration, and (or) temperatures) would likely affect the dominance order of STEC found in the present study. Different concentrations and combinations of multiple strains (more than two) should be examined to more realistically reflect the natural variation present in feces.
Further studies are necessary to fully determine the influence of environmental conditions including limiting nutrient concentrations on competitive ability of STEC with various stx and virulence gene profiles in comparison with stx-negative strains. A review of 90 STEC outbreaks from around the world, occurring between 1982 and 2006, found the vehicle of transmission was unknown in nearly 30% of outbreaks (Snedeker et al. 2009), suggesting that traceability of these pathogens is limited. Furthermore, the inability to detect certain serotypes may result in an underestimation of pathogenic non-O157 STEC. A culture bias could contribute to an underestimation of disease-associated serotypes and difficulty in traceback investigations if the pathogens are out-competed by other E. coli or bacteria during enrichment. Future studies should examine STEC culture bias during enrichment of environmental, as well as food and feces, samples, with diverse native microbiota, spiked with known concentrations of additional STEC.
The initial intent of this study was a proof of concept to examine competition causing potential bias between a mix of solely two (out of 200) different O-serogroups, strains with differing toxin gene profiles, but not considering any other (sub)type characteristics (e.g., H-type and (or) toxin subtype). For simplicity, only one enrichment media was used. However, no standard media exist for STEC detection and several different protocols are available each utilizing a different enrichment media. For example, the U.S. Department of Agriculture Food Safety and Inspection Service and the U.S. Food and Drug Administration procedures recommend tryptic soy broth and buffered peptone water, respectively. STEC enrichment bias using these broths has not been examined; however, we suspect that each enrichment medium could potentially produce biased cultures. Therefore, we suggest that future studies include a variety of media to further examine the level of potential STEC enrichment bias. Importantly, introduction of such a bias would critically influence outbreak trace-back investigations. Bias during outbreak investigations could have several outcomes, including the relevant pathogen being missed and the transmission vehicle being unidentified, or outbreaks attributed to the wrong pathogen/strain due to competition during enrichment (Pettengill et al. 2012). With continued emergence of novel pathogens, advances in STEC detection can potentially improve food safety monitoring systems, decreasing circulation of compromised food products and minimizing risks to human health.
Acknowledgements
The authors would like to thank Susanne Trapp and Yidong Han for their assistance in the laboratory. We thank Shaun Cook for expertise and valuable discussion. This research was supported by a grant from Alberta Innovates BioSolutions.
References
Akerlund T, Nordström K, and Bernander R. 1995. Analysis of cell size and DNA content in exponentially growing and stationary-phase batch cultures of Escherichia coli. Journal of Bacteriology, 177(23): 6791–6797.
Bai J, Paddock ZD, Shi X, Li S, An B, and Nagaraja TG. 2012. Applicability of a multiplex PCR to detect the seven major Shiga toxin-producing Escherichia coli based on genes that code for serogroup-specific O-antigens and major virulence factors in cattle feces. Foodborne Pathogens and Disease, 9(6): 541–548.
Bettelheim KA. 2007. The non-O157 Shiga-toxigenic (verocytotoxigenic) Escherichia coli; under-rated pathogens. Critical Reviews in Microbiology, 33(1): 67–87.
Boye E, and Lobner-Olesen A. 1991. Bacterial growth control studied by flow cytometry. Research in Microbiology, 142(2–3): 131–135.
Carter MQ, Brandl MT, Louie JW, Kyle JL, Carychao DK, Cooley MB, Parker CT, Bates AH, and Mandrell RE. 2011. Distinct acid resistance and survival fitness displayed by Curli variants of enterohemorrhagic Escherichia coli O157:H7. Applied and Environmental Microbiology, 77(11): 3685–3695.
Conrad CC, Gilroyed BH, McAllister TA, and Reuter T. 2012. Synthesis of O-serogroup specific positive controls and real-time PCR standards for nine clinically relevant non-O157 STECs. Journal of Microbiological Methods, 91(1): 52–56.
Conrad CC, Stanford K, McAllister TA, Thomas J, and Reuter T. 2014. Further development of sample preparation and detection methods for O157 and the top 6 non-O157 STEC serogroups in cattle feces. Journal of Microbiological Methods, 105(1): 22–30.
Cooley MB, Jay-Russell M, Atwill ER, Carychao D, Nguyen K, Quiñones B, Patel R, Walker S, Swimley M, Pierre-Jerome E, Gordus AG, and Mandrell RE. 2013. Development of a robust method for isolation of Shiga toxin-positive Escherichia coli (STEC) from fecal, plant, soil and water samples from a leafy greens production region in California. PLoS One, 8(6): e65716.
Dunbar J, White S, and Forney L. 1997. Genetic diversity through the looking glass: effect of enrichment bias. Applied and Environmental Microbiology, 63(4): 1326–1331.
Durso LM. 2013. Primary isolation of Shiga toxigenic Escherichia coli from environmental sources. Journal of Environmental Quality, 42(5): 1295–1307.
Eydallin G, Ryall B, Maharjan R, and Ferenci T. 2014. The nature of laboratory domestication changes in freshly isolated Escherichia coli strains. Environmental Microbiology, 16(3): 813–828.
Franz E, Delaquis P, Morabito S, Beutin L, Gobius K, Rasko DA, Bono J, French N, Osek J, Lindstedt BA, Muniesa M, Manning S, LeJeune J, Callaway T, Beatson S, Eppinger M, Dallman T, Forbes KJ, Aarts H, Pearl DL, Gannon VP, Laing CR, and Strachan NJ. 2014. Exploiting the explosion of information associated with whole genome sequencing to tackle Shiga toxin-producing Escherichia coli (STEC) in global food production systems. International Journal of Food Microbiology, 187(1): 57–72.
Gill A, and Gill CO. 2010. Non-O157 verotoxigenic Escherichia coli and beef: a Canadian perspective. Canadian Journal of Veterinary Research, 74(3): 161–169.
Gorski L. 2012. Selective enrichment media bias the types of Salmonella enterica strains isolated from mixed strain cultures and complex enrichment broths. PLoS One, 7(4): e34722.
Hussein HS. 2007. Prevalence and pathogenicity of Shiga toxin-producing Escherichia coli in beef cattle and their products. Journal of Animal Science, 85(13): E63–E72.
Kaspar C, Doyle ME, and Archer J. 2010. White paper on non-O157: H7 Shiga toxin-producing E. coli from meat and non-meat sources. Food Research Institute Food Safety Reviews [online]: Available from https://fri.wisc.edu/resources_food_reviews.php?brief=Foodborne+Illness [accessed 8 February 2016].
Lainhart W, Stolfa G, and Koudelka GB. 2009. Shiga toxin as a bacterial defense against a eukaryotic predator, Tetrahymena thermophila. Journal of Bacteriology, 191(16): 5116–5122.
Lin A, Sultan O, Lau HK, Wong E, Hartman G, and Lauzon CR. 2011. O serogroup specific real time PCR assays for the detection and identification of nine clinically relevant non-O157 STECs. Food Microbiology, 28(3): 478–483.
Liu B, Yin X, Feng Y, Chambers JR, Guo A, Gong J, Zhu J, and Gyles CL. 2010. Verotoxin 2 enhances adherence of enterohemorrhagic Escherichia coli O157: H7 to intestinal epithelial cells and expression of β1-integrin by IPEC-J2 cells. Applied and Environmental Microbiology, 76(13): 4461–4468.
Martínez-Castillo A, and Muniesa M. 2014. Implications of free Shiga toxin-converting bacteriophages occurring outside bacteria for the evolution and the detection of Shiga toxin-producing Escherichia coli. Frontiers in Cellular and Infection Microbiology, 4(1): 46.
Mathusa EC, Chen Y, Enache E, and Hontz L. 2010. Non-O157 Shiga toxin-producing Escherichia coli in foods. Journal of Food Protection, 73(9): 1721–1736.
Muniesa M, Blanch AR, Lucena F, and Jofre J. 2005. Bacteriophages may bias outcome of bacterial enrichment cultures. Applied and Environmental Microbiology, 71(8): 4269–4275.
Paddock Z, Shi X, Bai J, and Nagaraja TG. 2012. Applicability of a multiplex PCR to detect O26, O45, O103, O111, O121, O145, and O157 serogroups of Escherichia coli in cattle feces. Veterinary Microbiology, 156(3–4): 381–388.
Pettengill JB, McAvoy E, White JR, Allard M, Brown E, and Ottesen A. 2012. Using metagenomic analyses to estimate the consequences of enrichment bias for pathogen detection. BMC Research Notes, 5(1): 378.
Possé B, De Zutter L, Heyndrickx M, and Herman L. 2008. Novel differential and confirmation plating media for Shiga toxin-producing Escherichia coli serotypes O26, O103, O111, O145 and sorbitol-positive and -negative O157. FEMS Microbiology Letters, 282(1): 124–131.
Sidari R, and Caridi A. 2011. Methods for detecting enterohaemorrhagic Escherichia coli in food. Food Reviews International, 27(2): 134–153.
Singer RS, Mayer AE, Hanson TE, and Isaacson RE. 2009. Do microbial interactions and cultivation media decrease the accuracy of Salmonella surveillance systems and outbreak investigations? Journal of Food Protection, 72(4): 707–713.
Snedeker KG, Shaw DJ, Locking ME, and Prescott RJ. 2009. Primary and secondary cases in Escherichia coli O157 outbreaks: a statistical analysis. BMC Infectious Diseases, 9(1): 144.
Vimont A, Vernozy-Rozand C, and Delignette-Muller ML. 2006. Isolation of E. coli O157:H7 and non-O157 STEC in different matrices: review of the most commonly used enrichment protocols. Letters in Applied Microbiology, 42(2): 102–108.
Vimont A, Vernozy-Rozand C, Montet MP, Bavai C, Fremaux B, and Delignette-Muller ML. 2007. Growth of Shiga-toxin producing Escherichia coli (STEC) and bovine feces background microflora in various enrichment protocols. Veterinary Microbiology, 123(1–3): 274–281.
Information & Authors
Information
Published In

FACETS
Volume 1 • Number 1 • January 2017
Pages: 114 - 126
Editor: Elena P. Ivanova
History
Received: 9 February 2016
Accepted: 17 May 2016
Version of record online: 17 August 2016
Copyright
© 2016 Conrad et al. This work is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Data Availability Statement
All relevant data are within the paper.
Key Words
Sections
Subjects
Authors
Author Contributions
CCC and TR conceived and designed the study.
CCC performed the experiments/collected the data.
CCC, KS, and TR analyzed and interpreted the data.
KS, TAM, JT, and TR contributed resources.
All drafted or revised the manuscript.
Competing Interests
The authors have declared that no competing interests exist.
Metrics & Citations
Metrics
Other Metrics
Citations
Cite As
Cheyenne C. Conrad, Kim Stanford, Tim A. McAllister, James Thomas, and Tim Reuter. 2017. Competition during enrichment of pathogenic Escherichia coli may result in culture bias. FACETS.
1: 114-126.
https://doi.org/10.1139/facets-2016-0007
Export Citations
If you have the appropriate software installed, you can download article citation data to the citation manager of your choice. Simply select your manager software from the list below and click Download.
Cited by
1. Using Pathogenic Escherichia coli Type III Secreted Effectors espK and espV as Markers to Reduce the Risk of Potentially Enterohemorrhagic Shiga Toxin-Producing Escherichia coli in Beef
2. Optimized methods for the targeted surveillance of extended-spectrum beta-lactamase-producing
Escherichia coli
in human stool
3. Acquisition of plasmids from Shiga toxin-producing Escherichia coli strains had low or neutral fitness cost on commensal E. coli
4. Optimised methods for the targeted surveillance of extended-spectrum beta-lactamase producing
Escherichia coli
in human stool
5. A Novel Enrichment-Free, Low-Volume Filtration and Rapid Lysis Method (Elr) in Combination with Real-Time Pcr for Detection of Shiga Toxin-Producing Escherichia Coli (Stec) in Water
6. Escherichia coli O157:H7 strains in bovine carcasses and the impact on the animal production chain
7. Application of a Commercial Salmonella Real-Time PCR Assay for the Detection and Quantitation of Salmonella enterica in Poultry Ceca
8. Molecular detection of Shiga toxin-producing Escherichia coli (STEC) O157 in sheep, goats, cows and buffaloes
9. An Overview of Shiga-Toxin Producing
Escherichia coli
Carriage and Prevalence in the Ovine Meat Production Chain
10. Detection of
Ralstonia pseudosolanacearum
in drain water based on concentration, enrichment and the use of a duplex TaqMan PCR test
11. Shiga Toxin-Producing and Enteroaggregative Escherichia coli in Animal, Foods, and Humans: Pathogenicity Mechanisms, Detection Methods, and Epidemiology
12. Evaluation of denaturing gradient gel electrophoresis (DGGE) and next generation sequencing (NGS) in combination with enrichment culture techniques to identify bacteria in commercial microbial-based products
13. Competition among Escherichia coli Strains for Space and Resources
14. Variability in Characterizing Escherichia coli from Cattle Feces: A Cautionary Tale
15. Multi-Year Persistence of Verotoxigenic Escherichia coli (VTEC) in a Closed Canadian Beef Herd: A Cohort Study
16. Air-Dried Brown Seaweed,
Ascophyllum nodosum
, Alters the Rumen Microbiome in a Manner That Changes Rumen Fermentation Profiles and Lowers the Prevalence of Foodborne Pathogens
17. Shiga‐toxin Producing
Escherichia coli
: Pathogenicity, Supershedding, Diagnostic Methods, Occurrence, and Foodborne Outbreaks
18. Limitations of Immunomagnetic Separation for Detection of the Top Seven Serogroups of Shiga Toxin–Producing Escherichia coli
