Introduction
Evolution can occur on different time scales. Frequently, however, we aim to infer evolutionary processes by looking back thousands of years rather than looking at the first steps of evolution. Integrating genotype-phenotype information across timescales can help us elucidate how evolution works (
Linnen and Hoekstra 2009). Contemporary evolution is defined as recent and ongoing evolutionary change occurring on an ecological timescale in wild populations (
Hendry and Kinnison 1999;
Hairston Jr. et al. 2005), ranging from a few generations to a few centuries. Characterizing contemporary evolution is relevant for conservation biology, and it is becoming increasingly important to understand as climate change accelerates rapid environmental change (
Stockwell et al. 2003). Several examples of contemporary evolution have been identified in nature (e.g., Lake Malawi cichlids,
Streelman et al. 2004; saltmarsh beetles,
Van Belleghem et al. 2018;
Heliconius butterflies,
Reed et al. 2011). These exemplars provide key information on the mode and pace of adaptation as we seek to understand how rapid evolution occurs. However, evolutionary processes, including the strength of selection and the pace and probability of evolution, vary greatly within and among taxa (
Kingsolver et al. 2001;
Siepielski et al. 2009;
Sanderson et al. 2022), illustrating the need for studies in well-established systems to better test hypotheses regarding evolutionary patterns.
Disentangling the effects of natural selection from other evolutionary mechanisms, such as genetic drift, is an important first step as we seek to determine what selective pressures organisms face. This clarification is critical in predicting how organisms will respond to environmental change. Parallel evolution, the repeated evolution of traits in related lineages in response to similar environmental conditions (
Simpson 1953), has historically been used as strong supporting evidence for natural selection (
Endler 1986;
Schluter and Nagel 1995;
Schluter et al. 2004;
Wake et al. 2011;
Bolnick et al. 2018). Consistent and directional changes in traits described as parallel evolution are indicative of natural selection because it is highly unlikely that these repeated patterns would emerge based on chance alone (
Endler 1986;
Schluter and Nagel 1995;
Schluter et al. 2004;
Wake et al. 2011). Identifying cases of parallel evolution occurring in contemporary time frames may thus provide insight into processes that allow organisms to adapt to rapidly changing selective regimes.
Parallel evolution across populations of threespine stickleback (
Gasterosteus aculeatus) that move into freshwater habitats is an iconic example of adaptation (e.g.,
Bell and Foster 1994;
Schluter et al. 2004;
Colosimo et al. 2005). The threespine stickleback is a small ancestrally marine fish with a Holarctic distribution that is characterized by bony armour consisting of lateral plates, dorsal plates, three dorsal spines, and two pelvic spines (
Hagen and Gilbertson 1973;
Reimchen 1983;
Bell and Foster 1994;
Barrett et al. 2010;
Bjærke et al. 2010). Populations that colonized freshwater environments have consistently diverged in a variety of traits related to morphology, physiology, behaviour, genetics, and life history when compared to their contemporary marine ancestors (
Bell and Foster 1994;
McKinnon 2002;
Barrett et al. 2010;
Jones et al. 2012). Freshwater threespine stickleback typically have a more streamlined body shape, reduced body armour (bony plates, spines, and pelvic girdle), decreased salt tolerance, differences in mate preference, dietary changes, and reduced fecundity (
Bell and Foster 1994;
McKinnon 2002). The reduction in the number of lateral plates and the role of selection in freshwater populations have been particularly well studied (e.g.,
Klepaker 1993;
Reimchen 2000;
Bell 2001;
Schluter et al. 2004;
Colosimo et al. 2005;
Barrett et al. 2008;
Leinonen et al. 2011b;
Marchinko et al. 2014). Parallel evolution favoring a reduction in the number of lateral plates in freshwater colonizing threespine stickleback populations is so widespread that it is almost certainly an adaptive response to selection and highly unlikely to be the sole result of other mechanisms such as genetic drift or ecological vicariance (
Schluter et al. 2004;
Roesti et al. 2015). Threespine stickleback thus provide a powerful model for investigating questions pertaining to contemporary evolution because they have a well-characterized ecology (
Hagen and Gilbertson 1973;
Bell and Foster 1994;
McKinnon 2002), exhibit variation linked to environmental conditions (
Colosimo et al. 2005;
Barrett et al. 2010), and a large number of genomic resources are available for the species (e.g.,
Colosimo et al. 2004;
Jones et al. 2012;
Rogers et al. 2012;
Glazer et al. 2015;
Peichel and Marques 2017;
Peichel et al. 2017).
Lateral plate phenotype in threespine stickleback is under the genetic control of a large-effect gene,
Ectodysplasin (
Eda), that singlehandedly explains over 75% of the variation in lateral plate phenotype (
Colosimo et al. 2004,
2005;
Cresko et al. 2004;
Peichel and Marques 2017). Recent work has also shown a key role for this gene's receptor (
Edar;
Laurentino et al. 2022). The completely-plated morphs (>30 plates,
Hagen and Gilbertson 1972) that dominate in Eastern Pacific marine populations are usually homozygous for the complete-plated
Eda allele (
Colosimo et al. 2005;
Barrett et al. 2008; although we note this is not necessarily the case elsewhere; see, for example,
DeFaveri and Merilä 2013). Likewise, the low-plated morphs (<10 plates;
Hagen and Gilbertson 1972) that often dominate in freshwater populations are homozygous for the low-plated
Eda allele, resulting in many freshwater populations at or near fixation for the low-plated
Eda allele (
Colosimo et al. 2005). Heterozygotes for
Eda display either complete or partial morphs (10–30 plates;
Hagen and Gilbertson 1972) and are rare in both marine and freshwater environments (
Colosimo et al. 2004;
Cresko et al. 2004).
Eda is also known to have pleiotropic effects and has been previously linked to differences in shape, which could help explain why lateral plate phenotype and shape vary together in freshwater adaptation (
Albert et al. 2008;
Barrett et al. 2009;
Aguirre and Bell 2012;
Rogers et al. 2012). Multiple studies have found the low-plated allele is present as standing genetic variation at a frequency of ∼1% in marine populations off the coast of British Columbia, Canada (
Colosimo et al. 2005;
Barrett et al. 2008;
Morris et al. 2018). This standing genetic variation gives populations the potential to rapidly adapt to new freshwater environments (
Colosimo et al. 2005;
Mäkinen et al. 2006) and provides a clear link between genotype and phenotype (
Colosimo et al. 2004;
Peichel and Marques 2017).
There exist, to our knowledge, only three described cases of contemporary evolution in threespine stickleback, and both the mechanisms responsible and the pace of evolution remain relatively poorly understood. In the first well-documented case, a pond in Bergen, Norway, was disconnected from the ocean in 1960, resulting in the rapid isolation of a population of marine stickleback from their previous marine habitat (
Klepaker 1993). This population was sampled for nine years and showed a progressive decrease in complete morphs, reduction in spine length, change in body form, and enlargement of the eyes over this time period. Similarly, threespine stickleback in an Icelandic fjord that was isolated from the ocean for a period of 12 years exhibited a reduced number of lateral plates, shorter pelvic spines, and variation in overall shape morphology compared to nearby marine populations (
Kristjánsson et al. 2002). In Loberg Lake, Alaska, the stickleback population was wiped out by rotenone poisoning in 1983 (
Bell et al. 2004). The lake was later recolonized by marine stickleback and annual sampling from 1990 to 2001 revealed a shift in the population from 96% complete morphs to 11% complete morphs (
Bell et al. 2004). Although these studies suggest that threespine stickleback rapidly respond to environmental changes related to isolation in freshwater, more replicates, as well as more comprehensive experiments and analyses, are needed to support initial findings on the pace and progression of adaptation observed early in the adaptive process. Hypotheses around factors driving freshwater adaptation include selection on osmoregulatory genes linked to
Eda (
Heuts 1947), increased cost of plate production due to calcium limitation in freshwater (
Giles 1983), a juvenile growth advantage for low morphs in freshwater leading to higher overwinter survival (
Marchinko and Schluter 2007;
Barrett et al. 2009), differences in predation regimes (
Reimchen 1992;
Reimchen 2000;
Leinonen et al. 2011b), and differences in swimming ability (
Reimchen 1992;
Reimchen 1994;
Bergstrom 2002).
Here we test a case of contemporary evolution in a recently identified population of marine threespine stickleback that has been isolated to Lost Lagoon, British Columbia, Canada, for nearly 100 years. The population was isolated from the surrounding ocean waters of Coal Harbour in 1916, and marine water input via pumps ceased in 1929 (
Carl 1932). Since this time, Lost Lagoon has become a freshwater environment. We created genetic and phenotypic profiles of Lost Lagoon fish and compared these to genetic and phenotypic profiles of nearby marine and freshwater populations to test the hypothesis that this system represents an example of contemporary evolution. We predicted that Lost Lagoon fish would be intermediate in both genetic and phenotypic terms when compared to nearby marine and freshwater populations, indicating short-term effects of adaptive change in response to freshwater.
Discussion
Results of this study support the hypothesis that threespine stickleback in Lost Lagoon have begun to undergo both genetic and phenotypic changes characteristic of adaptation to freshwater in the nearly 100 years since the population was isolated. Our results support our prediction that Lost Lagoon stickleback are genetically and phenotypically intermediate but are following a parallel trajectory to that described by the shape differences between marine and established freshwater groups. While our data do not allow us to exclude the possible confounding effects of environmental effects or genetic drift on the phenotypes we observe, our results suggest that the Lost Lagoon population has been captured in the midst of this evolutionary transition.
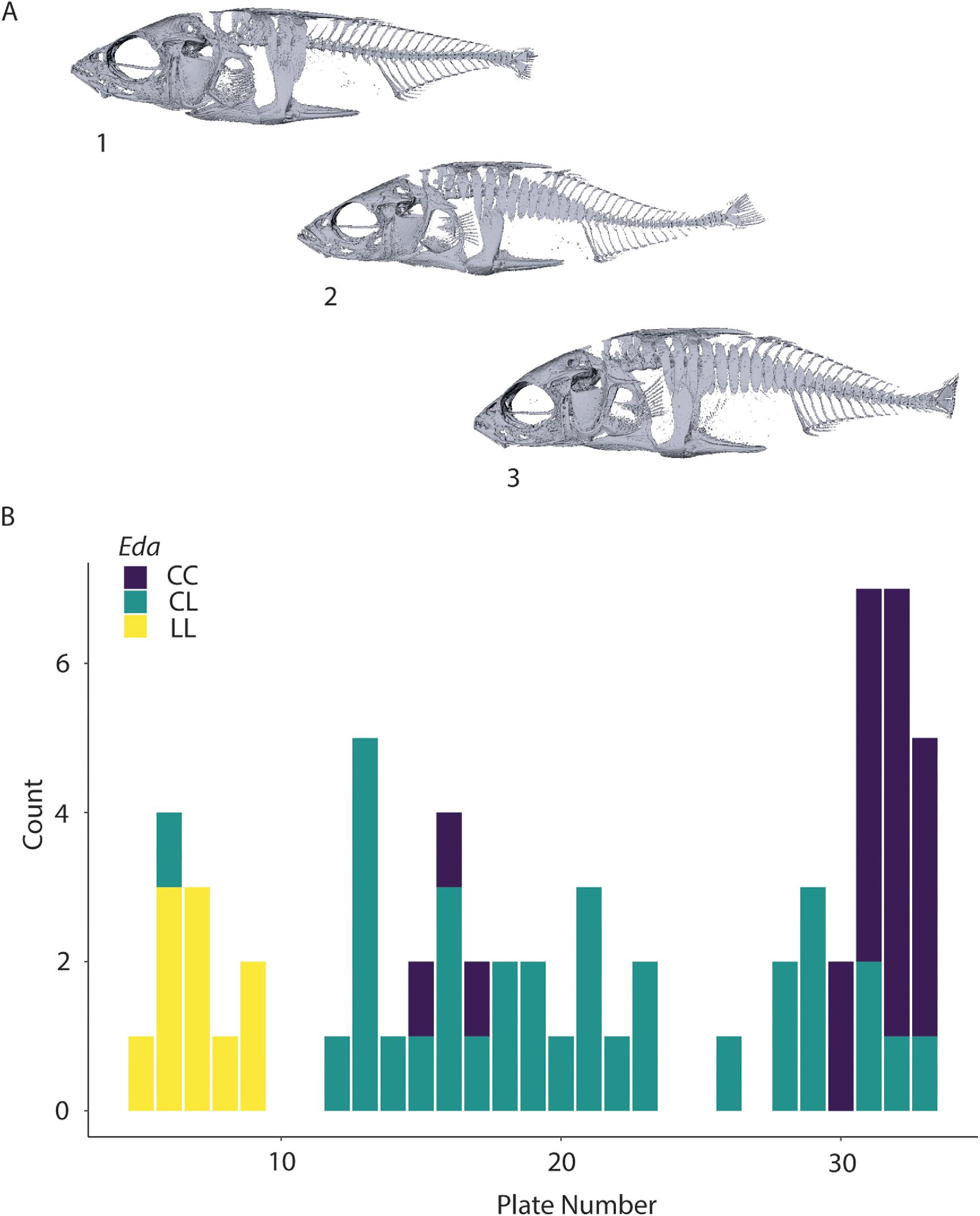
All three
Eda genotypes are present within the Lost Lagoon threespine stickleback population, and these genotypes are reflected in lateral plate morph as expected (
Fig. 2;
Colosimo et al. 2004;
Cresko et al. 2004;
O'Brown et al. 2015;
Peichel and Marques 2017). The tight match between
Eda genotypes and lateral plate morphs provides further support for the well-established claim that
Eda acts as a large effect gene and explains greater than 75% of the variation in the number of lateral plates (
Colosimo et al. 2004).
O'Brown et al. (2015) found that low-plated
Eda alleles induce
cis-acting regulatory changes, resulting in lower levels of expression in developing skeletal features such as lateral plates and spines compared to the complete plated
Eda allele. The difference in expression proposed by
O'Brown et al. (2015) provides a molecular mechanism that links
Eda genotype and lateral plate morphs consistent with what is observed across the Holarctic (
Colosimo et al. 2004;
Cresko et al. 2004;
Peichel and Marques 2017) and mirrored in Lost Lagoon.
Previous studies have linked
Eda to variation in body shape, beyond lateral plate phenotype, through quantitative trait locus analysis (
Albert et al. 2008;
Rogers et al. 2012). Our results support this link in Lost Lagoon (
Table 1). Although
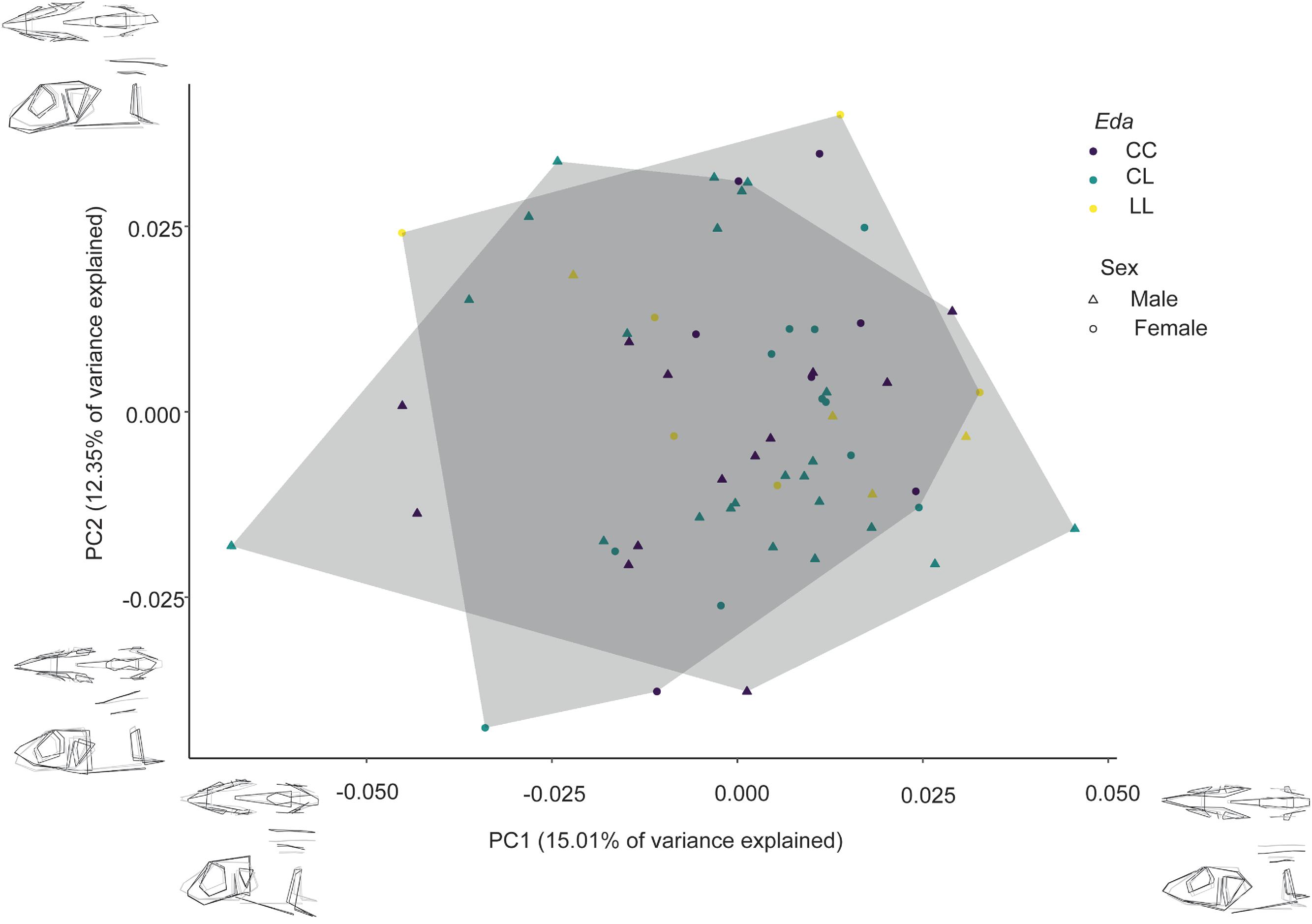
Aguirre and Bell (2012) observed a significant association between body shape and lateral plate morph, they noted that the mean variation in body shape between morphs was small and mostly associated with the size of bones. The divergence of homozygous low-plated individuals along PC2 in this study (
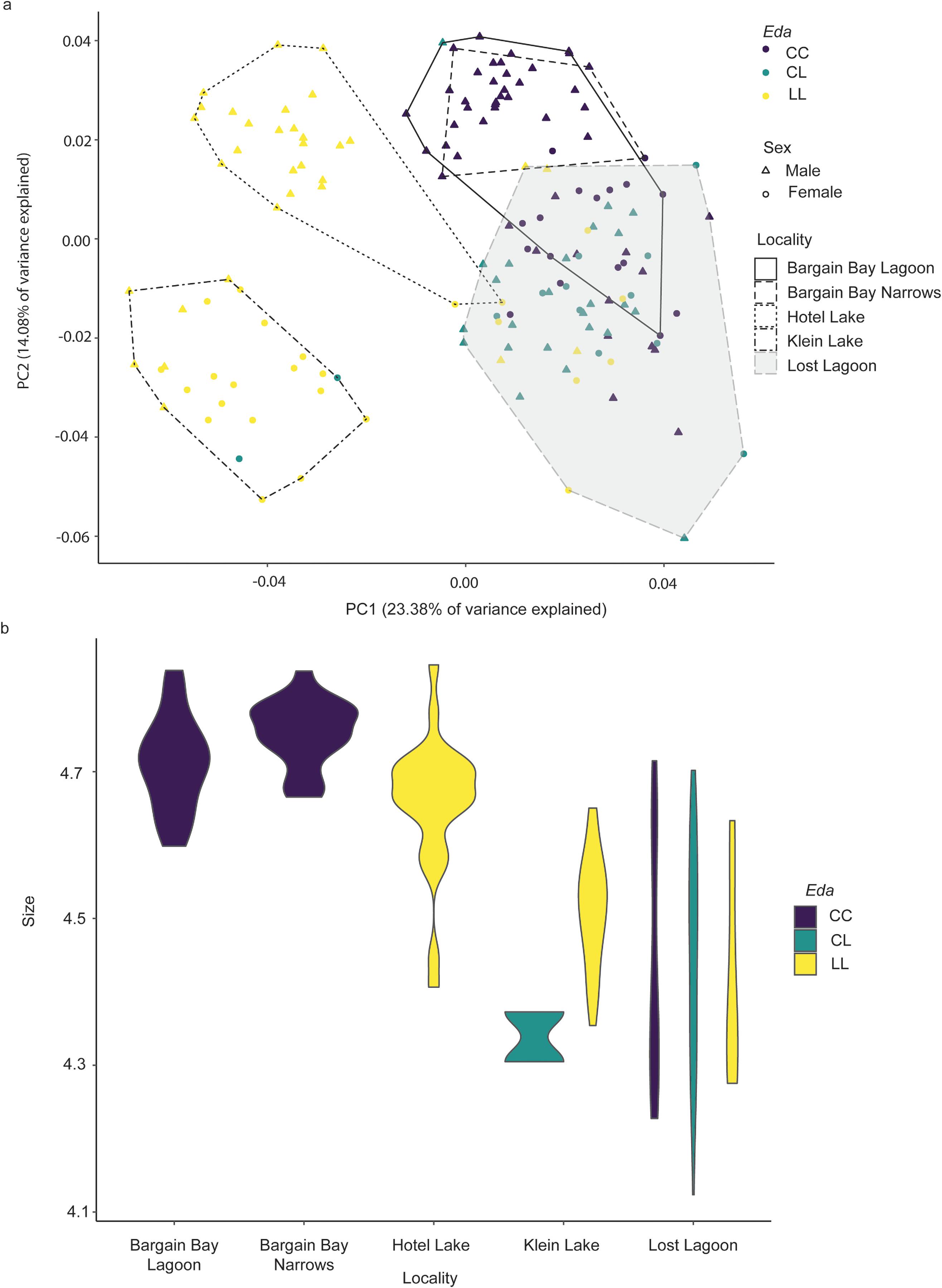
Fig. 3) provides support for previous studies that suggest
Eda has pleiotropic effects on shape (
Albert et al. 2008;
Barrett et al. 2009;
Rogers et al. 2012) and is consistent with the observation that the number of lateral plates and shape vary in the same direction during freshwater adaptation (
Klepaker 1993;
Bell and Foster 1994;
Schluter et al. 2004;
Bjærke et al. 2010;
Aguirre and Bell 2012).
Eda heterozygotes and complete-plated homozygotes diverge very slightly in shape in the Lost Lagoon sample, and unsurprisingly, heterozygotes occupy more morphospace, indicating greater within-group variation.
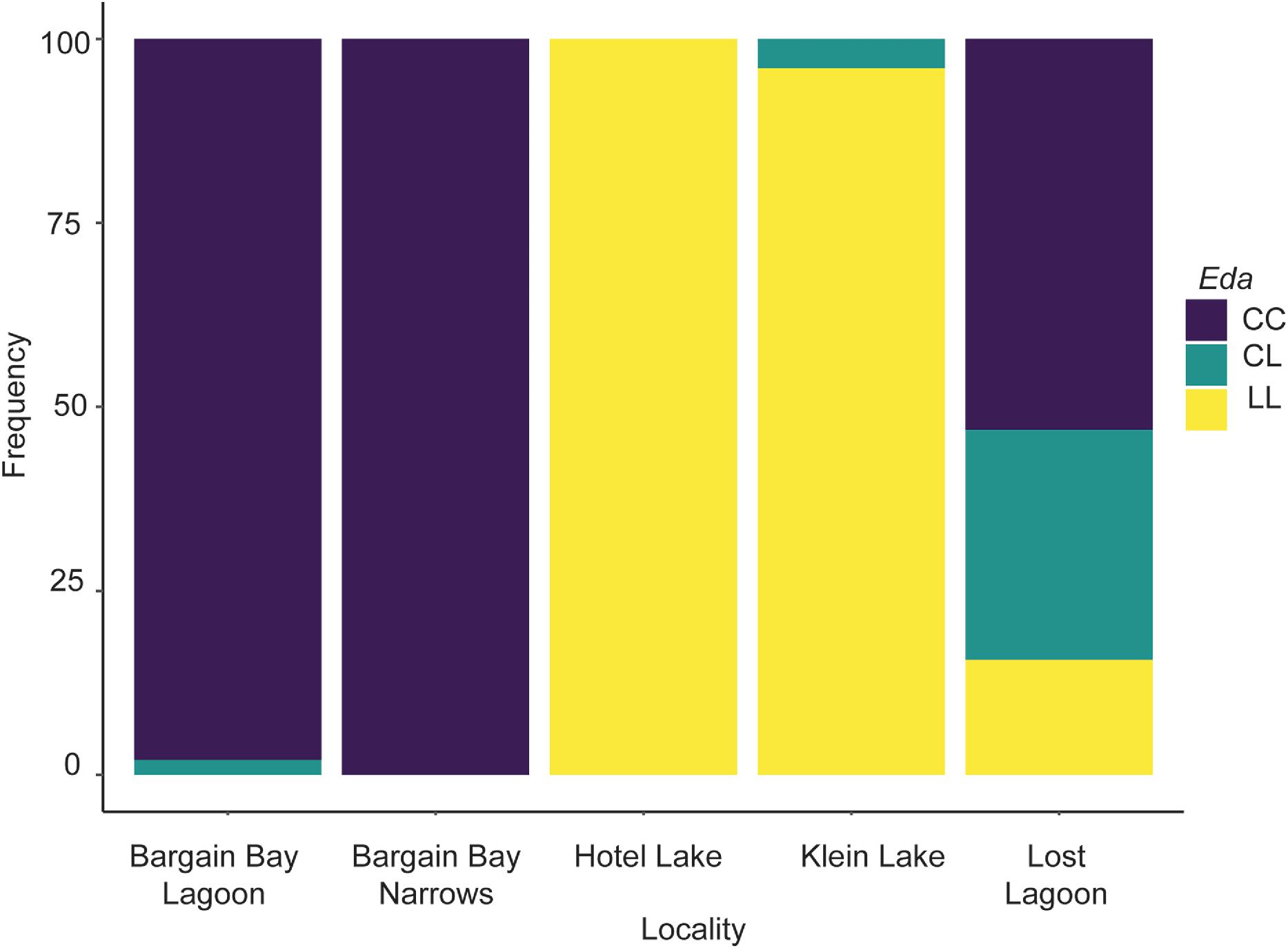
When compared to nearby marine populations in Madeira Park, threespine stickleback in Lost Lagoon appear to have begun to adapt to freshwater conditions, supporting the main hypothesis for this study. The first and most obvious indication of adaptation is that the frequency of the low-plated
Eda allele, and corresponding lateral plate phenotype, is much higher in Lost Lagoon at 42.19% than in the neighbouring marine populations of Bargain Bay Lagoon and Bargain Bay Narrows (
Fig. 4). The low-plated
Eda allele is absent in the sample from Bargain Bay Narrows and present at a frequency of 1% in Bargain Bay Lagoon (
Morris et al. 2018), confirming that the complete-plated allele dominates marine environments off the coast of British Columbia, Canada. The presence of the low-plated allele in Bargain Bay Lagoon supports previous findings that the low-plated allele is present in marine populations as standing genetic variation (
Colosimo et al. 2005;
Barrett and Schluter 2008;
Morris et al. 2018). The lack of low-plated
Eda alleles in Bargain Bay Narrows is likely an artifact of the relatively small sample size (
n = 50). Although the frequency of the low-plated
Eda allele in Lost Lagoon is decidedly elevated compared to nearby marine populations, it is not nearly as high as in nearby freshwater populations in Madeira Park, Hotel and Klein Lakes, which were presumably colonized following deglaciation thousands of years ago (
Fig. 4).
Pairwise
FST, calculated using nongenic SNPs, provides further support for the hypothesis that Lost Lagoon is in a transitional state, beginning to diverge from marine populations but not yet as divergent as well-established freshwater populations. Results indicate Lost Lagoon is much more genetically differentiated from nearby marine populations than marine populations are from one another, indicating Lost Lagoon has genetically diverged from its presumably ancestral condition over the last ∼100 years (
Table 2). However, Lost Lagoon remains more genetically similar to marine populations than the established freshwater populations to which it was compared (
Table 2). Freshwater populations are as differentiated from one another as they are from marine populations at neutral loci due to isolation. While these results do not rule out the possibility that genetic differentiation in Lost Lagoon is the result of gene flow between marine and freshwater populations, this is unlikely based on the geography of the area. There are two potential seasonal inflows into Lost Lagoon: Ceperley Creek, which does not connect to a natural water reservoir but rather pumps municipal water into Lost Lagoon; and Beaver Lake, which may occasionally overflow in times of very high water levels. We have recently (2023) confirmed the presence of
G. aculeatus in Beaver Lake. Passive dispersal caused by human activities, either intentionally or unintentionally, is also possible, especially in such an urban setting. Additionally, birds are commonly believed to be capable of moving fish eggs, although the empirical evidence for this phenomenon in stickleback is scant (
Hirsch et al. 2018). Nonetheless, freshwater stickleback may contribute to the Lost Lagoon population.
Working with pooled data raises concerns about adequate sample size for accurate
FST estimates, as large sample sizes have previously been viewed as essential. However,
Willing et al. (2012) found that very small sample sizes can obtain accurate
FST estimates when the number of genetic markers is large in simulated data, and this result has since been replicated in sequenced data (
Nazareno et al. 2017). Using pooled sequencing results and an appropriate
FST estimator that considers differences between pooled data and individual data produces accurate results when compared to individual sequencing results (
Gautier et al. 2013;
Hivert et al. 2018). The
FST estimator in Poolfstat based on the analysis of variance that was developed and tested by
Hivert et al. (2018) is unbiased and measures
FST with accuracy when compared to results from individual sequencing as well as previous estimators for pooled sequencing. Pairwise
FST values calculated in this study using Poolfstat with over four million SNPs can be reasonably interpreted as accurate based on these findings. Further analysis can be conducted to determine specific components of the population structure.
The results of morphometric analyses comparing shape among all individuals from all localities revealed significant effects of size, sex, and
Eda on body shape as well as an interaction between size and
Eda (
Table 3), confirming the presence of nonuniform effects of size on shape in these samples. In particular, individuals from Lost Lagoon display a range of body sizes that most closely resemble those found in the freshwater localities, but rather than clustering with those localities as one would expect if PC1 were a “size component”, instead Lost Lagoon individuals cluster with the considerably larger marine individuals along this axis. This indicates that PC1 is relatively more heavily influenced by
Eda genotype than size (
Fig. 5). Divergence of marine and freshwater body shape in individuals from Madeira Park supports many previous studies that found parallel evolution in body shape varies in the same direction as lateral plate phenotype (
Klepaker 1993;
Bell and Foster 1994;
Bell 2001;
Schluter et al. 2004;
Bjærke et al. 2010;
Aguirre and Bell 2012).
Interestingly, Lost Lagoon stickleback resemble marine individuals in overall body shape, regardless of
Eda genotype and corresponding lateral plate phenotype, despite their relatively smaller size. This suggests that Lost Lagoon stickleback are more similar in shape to marine populations characterized by a deeper body, a larger pelvic girdle, larger dorsal plates, a shorter head and blunter snout, and larger eyes (
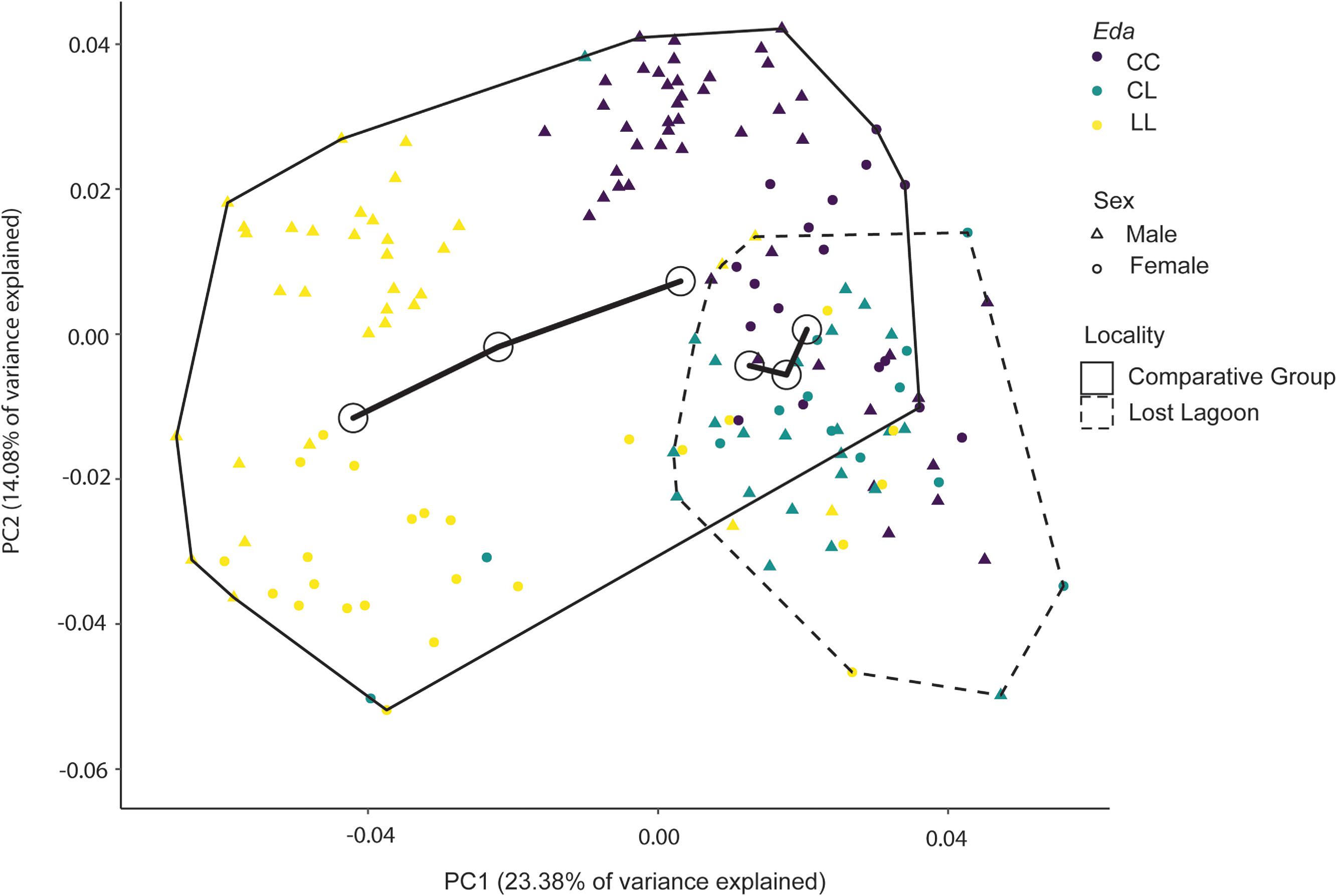
Fig. 5). Lost Lagoon stickleback diverge at least partially from marine populations genetically and in lateral plate phenotype, yet they cluster with marine populations along the main axis of variation in shape. Interestingly, phenotypic trajectory analysis does not indicate that the shape trajectory calculated for Lost Lagoon differs from that calculated for the comparison group, despite the presence of a relatively highly variable intermediate phenotype that clusters separately from most other individuals in the comparison group (
Fig. 6). One possible explanation for this discrepancy is that the pace of adaptation differs between different phenotypic characteristics.
Schluter et al. (2004) proposed that parallel evolution of the lateral plate phenotype relies on only a few genes with large effects, namely
Eda, whereas parallel evolution of body shape relies on many genes with smaller effects. The results we report here support this hypothesis, although we note that our failure to observe significant differences in trajectory features is not a direct test of the similarity of these trajectories.
Lost Lagoon individuals cluster apart from marine individuals slightly along the second principal component. Individuals at either end of this axis vary in bone size, with Lost Lagoon stickleback trending towards exhibiting a smaller pelvic girdle and smaller dorsal plates (
Fig. 5). However, this axis of variation may also be associated with sex. Male-dominated samples from Bargain Bay Narrows, Hotel Lake, and, to some extent, Bargain Bay Lagoon cluster at the positive end, while our female-dominated Klein Lake sample clusters near the negative end of the axis. Divergence of males towards the positive end and females towards the negative end of this axis also occurs within groups, except for Lost Lagoon, where males and females are interspersed. The most striking occurrence is in Hotel Lake, where the only two females present greatly diverge from the rest of the population. This divergence suggests that this axis of variation may be capturing variation that is partly driven by sex. Several previous studies have observed sexually mediated variation in morphological traits in stickleback (e.g.,
Aguirre et al. 2008;
Aguirre and Akinpelu 2010;
Leinonen et al. 2011a;
Kitano et al. 2012), including between individuals from the comparative localities described here (
Schutz et al. 2022). Interestingly, while there is a significant effect of sex on shape in Lost Lagoon (
Table 1), the nature of this variation is not clearly captured along either of the two largest principal components of variation within this group (
Fig. 3). Sexually mediated variation in stickleback is known to vary in expression across habitats (
Spoljaric and Reimchen 2008) and may be obscured by habitat-associated differences in body shape (
Schutz et al. 2022). In the Lost Lagoon population documented here, which is in the process of adapting to fresh water, changing
Eda allele frequencies are likely driving gradual body shape change that is obscuring the nature of any sexually mediated variation in body shape within the Lost Lagoon sample. We also note that single-locus sex determination may be less reliable than a multi-locus approach (
Toli et al. 2016), and while sexual dimorphism is not a central focus of this work, determining the relationship between sex and
Eda may require a more complex consideration of sex determination.
Our results provide an interesting comparison to the three other well-documented examples of contemporary evolution in stickleback described above (
Klepaker 1993;
Kristjánsson et al. 2002;
Bell et al. 2004). The time scale in these three cases is much shorter than in the present case, ranging from a maximum of ∼40 years in the Norwegian example (
Klepaker 1993) to ∼12 years in the Icelandic example (
Kristjánsson et al. 2002) to less than ten years in the Alaskan example (
Bell et al. 2004). However, in all of these cases, the decline in low-plated morphs was much more pronounced than what we report here, indicating that the pace of evolution in different genetic and environmental contexts may be quite different. While we do not have the data to test the hypothesis about pace in the present study, this discrepancy suggests that further characterization of the evolutionary context will be important as we seek to further understand adaptive change in sticklebacks. We also note that these differences reinforce the caveat we raised earlier in the Discussion, namely that our data do not allow us to exclude the influence of either drift or environmental effects on the phenotypes we observe.
Taken together, the results of this study indicate that Lost Lagoon stickleback appear to be in the process of adapting both genetically and phenotypically to a freshwater environment, likely following the same parallel evolutionary trajectory that is understood to be common among transitioning stickleback in Eastern Pacific habitats, and our data reflect a snapshot of this transition at an intermediate point. We demonstrate, for the first time, the direct effects of changing
Eda frequency on the lateral plate phenotype, providing an example of the genetic basis for parallel evolution on a contemporary timescale. In this example, the frequency of
Eda genotypes and lateral plate phenotypes is not consistent with those that are characteristic of either marine or fully freshwater populations. Further, although Lost Lagoon is genetically distinct from nearby marine populations, these fish still more closely resemble those marine populations in overall body shape relative to either freshwater comparator, despite occupying a unique area of morphospace relative to the comparative populations studied here, suggesting an intermediate phenotype along this evolutionary trajectory.
Eda frequency and lateral plate phenotype, as assessed in this study, seem to be changing faster than body shape in response to freshwater conditions, suggesting that future work might consider that the pace of adaptation differs across traits in response to the same environmental conditions in this system. Few stickleback populations have been identified in nature where all three
Eda genotypes and corresponding lateral plate morphs are present at reasonably high frequencies. In those that have been identified, polymorphism is typically maintained by a balance between selection and gene flow (
Raeymaekers et al. 2014) or disruptive natural selection (
Marchinko et al. 2014). While these populations may be useful in testing for differences between
Eda genotypes in a specific habitat, gene flow complicates the interpretation of results because individuals with different
Eda genotypes do not necessarily have a shared genetic background, as some may be recent migrants. The presence of all three
Eda genotypes in an isolated population in Lost Lagoon makes it an ideal population to test hypotheses for adaptation because of the natural presence of variation in
Eda on a shared genetic background without gene flow. The results presented in this study support the findings of previous studies demonstrating contemporary evolution in stickleback (
Klepaker 1993;
Kristjánsson et al. 2002;
Bell et al. 2004), and our results further bolster the case for
G. aculeatus as a key model to investigate the strength and temporal dynamics of selection (
Kingsolver et al. 2001;
Siepielski et al. 2009). Future studies of this population should include repeated sampling over multiple years to determine the rate of evolutionary change in this population (
Lande and Arnold 1983;
Hendry and Kinnison 1999;
Sanderson et al. 2022) to better compare with other known examples of contemporary evolution, including such systems as the Lake Malawi cichlids (
Streelman et al. 2004), saltmarsh beetles (
Van Belleghem et al. 2018),
Heliconius butterflies (
Reed et al. 2011), and others, which are powerful models to understand such evolutionary processes in detail.