Introduction
Agroecosystems contain a mosaic of crop and non-crop habitat, the balance of which is an important determinant of arthropod biodiversity as non-crop habitat provides a spatiotemporally stable source of food, shelter, nesting, and mating resources (
Dolezal et al. 2022). Specifically, variation in arthropod community composition has been linked to local attributes, including plant biomass, structural complexity, and plant community composition (
Stinson and Brown 1983;
Schaffers et al. 2008;
Borer et al. 2012;
Prather and Kaspari 2019) as well as landscape-level measures of habitat diversity, land-use intensity, landscape connectivity, and landscape complexity (
Schweiger et al. 2005;
Hendrickx et al. 2007;
Fahrig et al. 2011;
Gossner et al. 2016;
Seibold et al. 2019). Compositional differences in arthropod communities may also result from dispersal limitation and ecological drift, effects of which are often exacerbated by patch isolation and especially affect groups with limited dispersal ability (
Bell 2001;
Hubbell 2001;
Vellend 2010). This is significant since many arthropod taxa have rapid generation times and thus the influences of temporal and spatial processes on community divergence can be powerful even within a single growing season (
Kingsolver 1989;
Chown and Gaston 1999). Given the paucity of non-crop habitat in many agroecosystems, measures aimed at maintaining and restoring natural areas can be very effective in promoting arthropod biodiversity (
Paterson et al. 2019;
Dolezal et al. 2022). Several studies have demonstrated that restored habitat can lead to an increase in arthropod richness and abundance, often with positive effects on ecosystem services such as herbivore suppression, pollination, and predation services (
Albrecht et al. 2020;
Paterson et al. 2019;
Dolezal et al. 2022).
Most of these community-shaping processes have been investigated from a spatial perspective, but temporal factors can also play a central role (
Grøtan et al. 2012). Seasonality is particularly important in many systems, stemming from an interplay of species-specific responses to abiotic conditions, such as temperature and precipitation, biotic conditions, such as plant resource availability, and stochastic variation through time (
Wolda 1978,
1988;
Stinson and Brown 1983;
Grøtan et al. 2012;
Hatosy et al. 2013). Previous studies have found that the effect of habitat composition and the configuration of those habitats in the landscape on arthropod communities can vary across the growing season (
Bertrand et al. 2016) and that landscape composition can modulate phenological diversity (
Sydenham et al. 2014). Even in tropical systems with less pronounced seasonality compared to temperate regions, arthropod community composition in both natural forests and rubber plantations shows high seasonal turnover (
Beng et al. 2018). Arthropods living in agroecosystems experience both seasonal climatic variation and season-dependent management actions such as plowing, planting, and pesticide application (
Anderson et al. 2012;
Beckmann et al. 2019;
Hausmann et al. 2019).
Based on these previous studies, our primary goal was to evaluate the relative impact of local, landscape, and seasonal variables for explaining variation in arthropod community composition across a highly heterogeneous agroecosystem landscape that varies in the coverage of dedicated cropland and native habitats. Our secondary goal was to evaluate the utility of metabarcoding of Malaise trap samples as a simple and repeatable means of monitoring arthropod biodiversity. Our third goal was to establish an arthropod biodiversity benchmark that spans a wide gradient of land use types by which the effectiveness of land restoration activities on the ALUS farm network might be evaluated in future years.
On the basis of the extensive body of insect community research described above, we predicted a positive relationship between arthropod community dissimilarity and environmental distances. That is, larger differences in environmental conditions should generate more dissimilar communities for environmental variables representing weather, plant community attributes, or agricultural extent across the landscape. To account for possible effects of dispersal limitation, ecological drift, and/or unquantified spatiotemporally structured environmental variables, we incorporated a novel framework proposed by
Jabot et al. (2020) for linking spatial and temporal distances as covariates in analyses with environmental variables.
Discussion
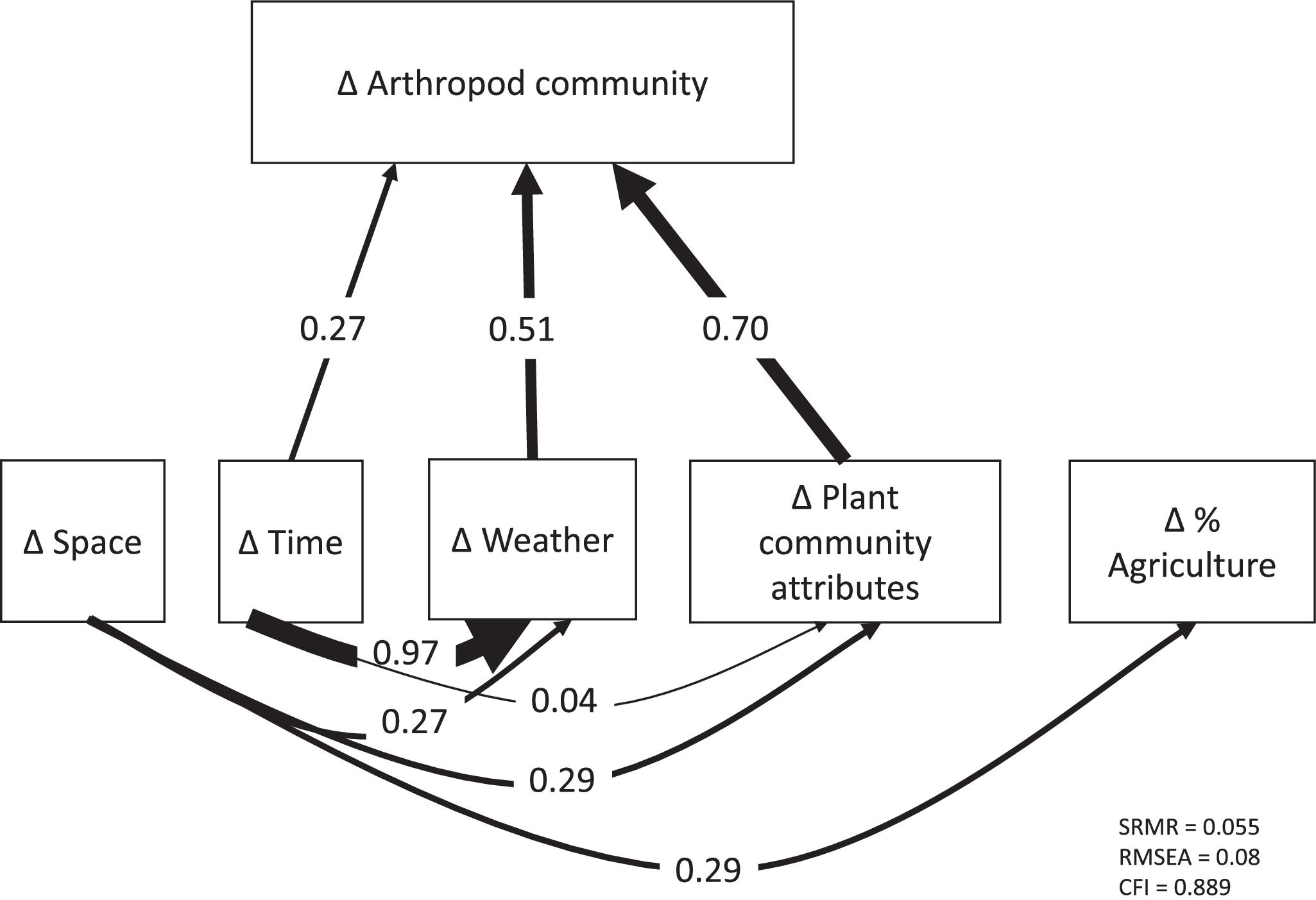
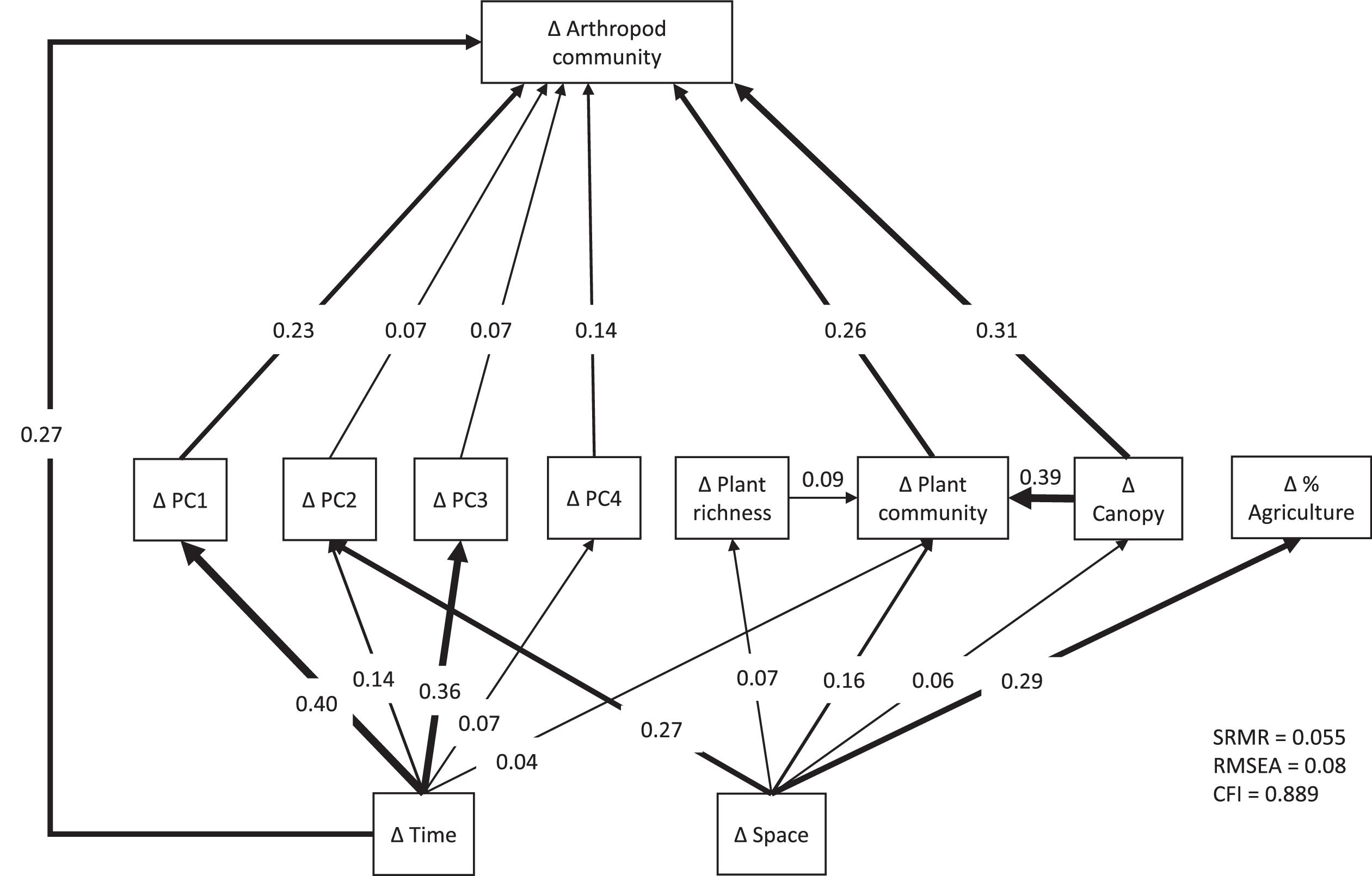
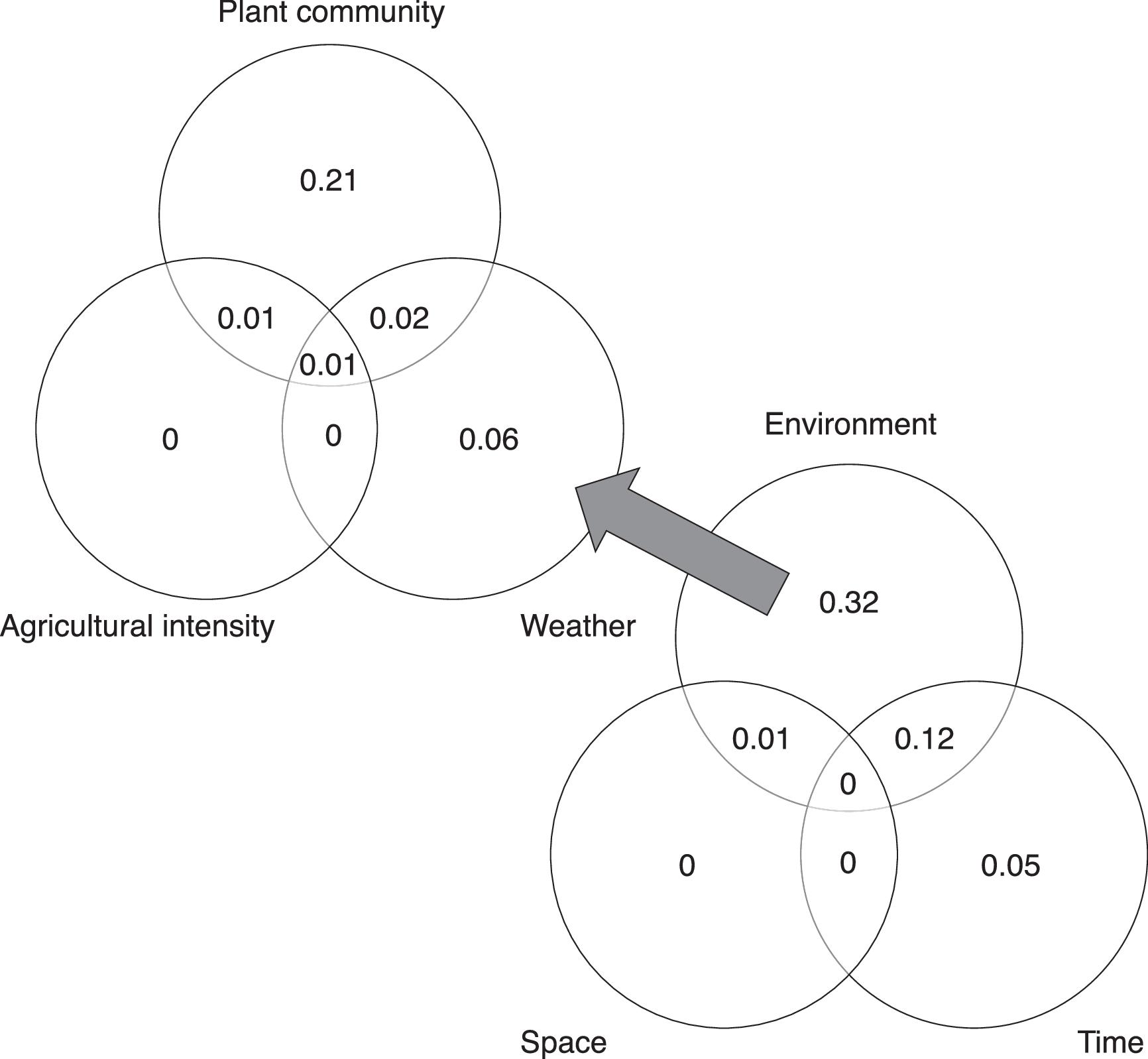
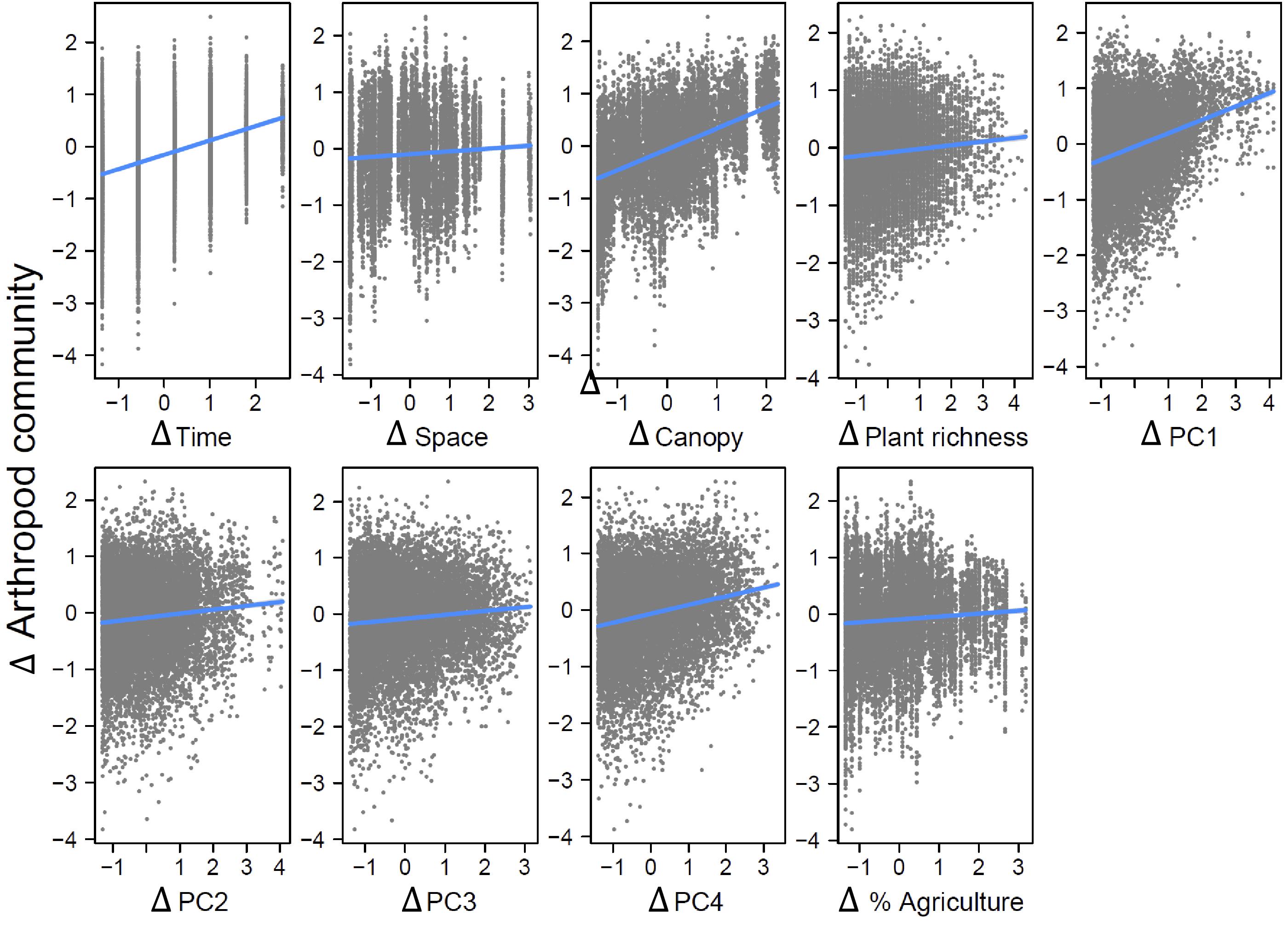
Our strongest finding was that spatial variation in arthropod community composition was largely shaped by spatial variation in local floristic composition and weather conditions. These effects dwarfed the impact of spatial variation in agricultural extent across local landscapes, for unknown reasons. We found arthropod communities to be highly variable among localities and across time periods. Plant community attributes best explained this variation. Variability in weather across the sampling season also played an important role. Environmental variables demonstrated both temporal and spatial structure and significant effects of temporal distance on arthropod dissimilarity remained even after accounting for environmental variables, while spatial distance did not retain a significant effect.
It is conceivable that the weak influence of agricultural extent was an artifact of confounded effects among agricultural extent across the local landscape, plant composition, and distance (
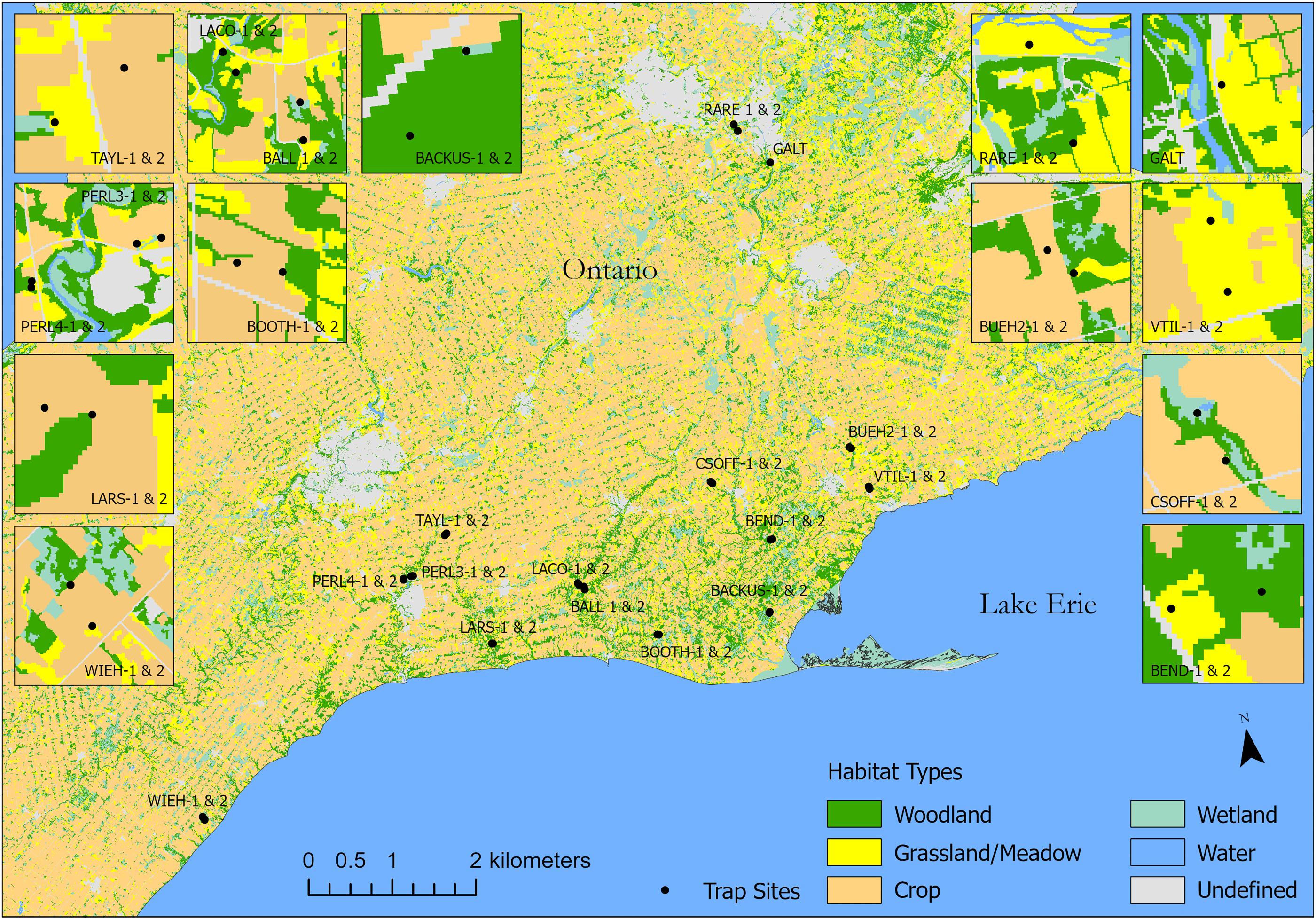
Fig. 1). Path analysis is designed specifically to accommodate cross-correlation in complex systems to reduce the possibility of such confounding effects (
Shipley 2016). We nonetheless repeated the path analysis and variance partitioning after removing the plant composition data. While this alternative model structure resulted in a slightly stronger effect of agricultural extent, it still had much weaker influence on arthropod community dissimilarity than any of the other variables (Figs. S2–S3). Variation in agricultural extent was positively related to distance between our study sites (Fig. S4,
P < 0.001,
R2 = 0.083), so it is conceivable that some of the impact of agricultural extent has been incorrectly assigned to distance. Spatial distance was also a weak and statistically insignificant predictor of arthropod beta diversity across our study sites, however, so potential confounding of agricultural extent with distance would not alter our overall conclusion that local variation in plant composition and cover combined with weather variation had the strongest effects on arthropod beta diversity in our study system.
Given that our study landscape supported a mixture of croplands, prairie, wetland, and woodland habitat (
Fig. 1), our study sites were no doubt far more heterogeneous than one would normally encounter in regions dominated by industrial farming. Our index of agricultural extent (% cover within a 2 km radius) is rather simplistic, because many different agricultural characteristics contribute to intensification effects on flora and fauna (
Beckmann et al. 2019). Resolving this issue will probably require a properly balanced design that is focused directly on more highly detailed comparison between sites with widely differing farming practices.
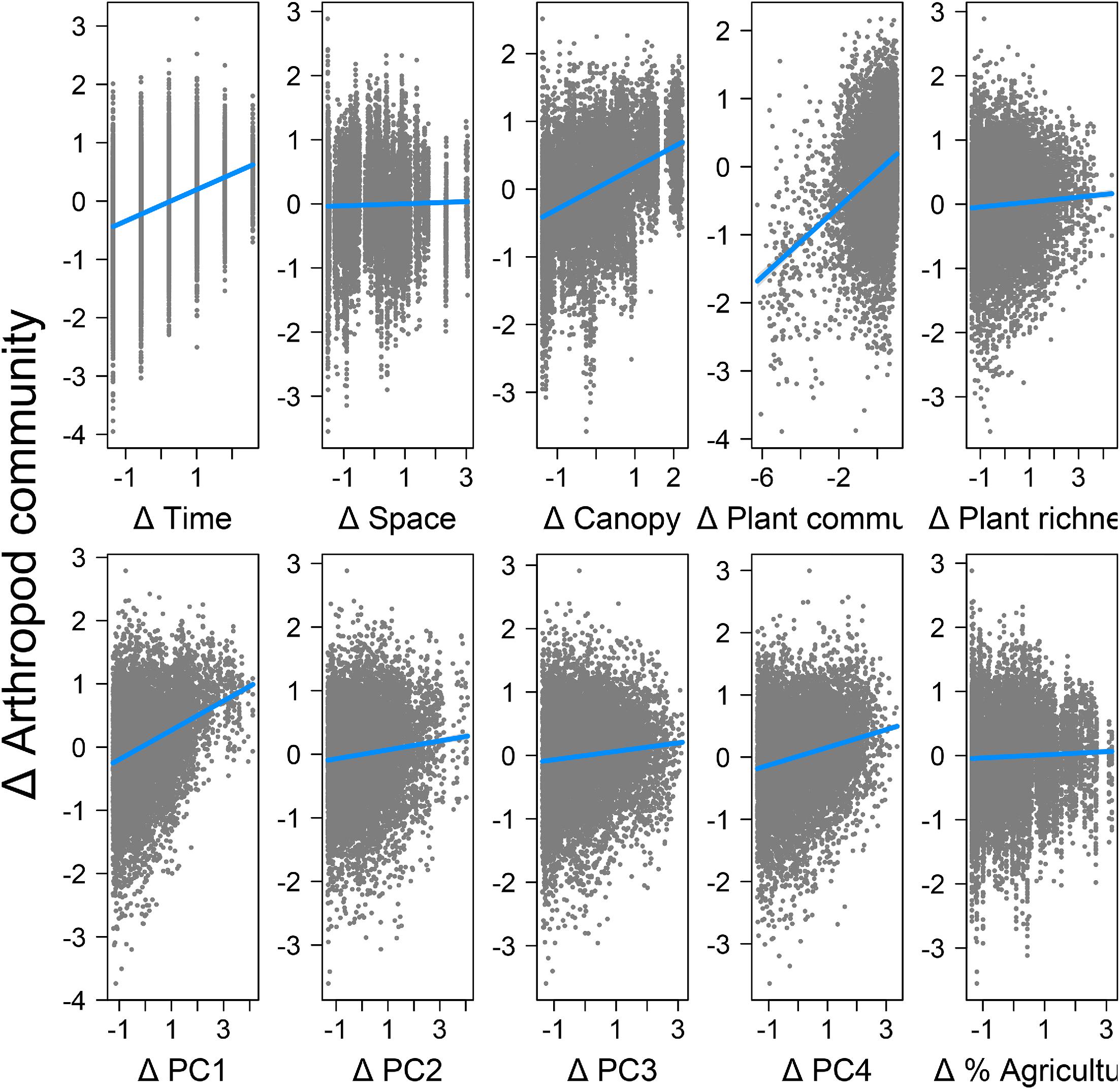
Canopy openness had a significant direct effect on differences in arthropod community composition, whereas plant richness did not, though both had indirect effects through plant community composition. It is noteworthy that plant richness alone did not have a significant direct effect in our analyses, as many studies have shown this to be an important determinant of arthropod community composition (
Borer et al. 2012;
Ebeling et al. 2018). Combined with the indirect effect through plant community composition, this means that the identities of plants mattered more than simple measures of richness for the arthropod communities studied here (e.g.,
Harvey and MacDougall 2015). This could be due to species-specific preferences for food (either directly on plants or other organisms that depend on those plants), nesting, shelter, and mating resources, or because plant community composition also acts as a reliable index of other environmental factors such as light availability or soil type (
Schaffers et al. 2008). If the functional and/or nutritional diversity of plant communities is low, it is thus likely that fewer arthropod taxa and functional groups will be represented in these communities (
Dolezal et al. 2022).
Taken together, these findings suggest that restoration of multiple habitat types with compositionally distinct plant communities at a local scale is likely to be an effective method for improving arthropod diversity in agroecosystems, provided that the landscape contains enough functionally connected habitat to maintain the species pool (
Scheper et al. 2013). Such an approach is exemplified well by
Windsor et al. (2021) via their use of network analyses to identify functionally complementary plant mixes that maximize arthropod biodiversity and ecosystem service provisioning in agroecosystems. Beta diversity enhancement through habitat modification is often promoted as one way of mitigating the biodiversity crisis, although it is a crude metric that can be enhanced as readily by addition of invasive species as by restoration of endemic species (
Socolor et al. 2016).
Variation in weather conditions, particularly across the growing season, also played an important role in determining arthropod community composition. This could be explained by several mechanisms. The first is that weather has a direct effect on arthropod survival and reproduction. Arthropods are generally constrained to narrow optimum ranges of temperature and humidity and taxa differ widely in their tolerances for climatic conditions, with some species being specialized for early emergence (
Høye and Forchhammer 2008). Such differences in the phenology of emergence due to weather conditions results in compositional turnover throughout the season. It could also partially explain why strong differences were observed with forest canopy, as turnover of arthropods between shaded cool forest and warmer and often drier herbaceous plant communities tends to be high (
Yekwayo et al. 2017). A related explanation could be resource limitation. Many arthropods depend on specific feeding and nesting/shelter resources, and many of those resources are not available early in the season due to plant phenology in the case of herbivores (foliage) and pollinators (flowers), and the phenology of prey in the case of predators (
Høye and Forchhammer 2008).
Much of the variation in arthropod community composition could be explained by environmental variables. The effect of spatial distance on community dissimilarity was not significant after accounting for environmental variation. This finding is especially interesting, given the large extent of our study region and degree of crop monoculture, the fact that many non-crop areas with natural or restored plant cover can be highly spatially isolated (
Fig. 1), and that this habitat isolation has been in place for many decades given that this region has been intensely farmed since at least the 1930 s (
Riley 2013;
McQuarrie 2014). This degree of habitat transformation over the last century might imply acute species turnover by spatial distance but this was not the case.
That being said, single individuals of many flying arthropods such as some species of bee have foraging ranges upwards of 5 km (
Greenleaf et al. 2007) and are likely to travel much further in windy conditions (
Pasek 1988), resulting in many transient individuals being caught in the traps and high dispersal potential. Indeed, one might expect an arthropod community dominated by highly mobile species in a region that has been repeatedly glaciated in the past. As well, the introduction of dietary and/or eDNA could lead to BIN identifications that are not representative of the immediate trap vicinity (
Braukmann et al. 2019). This DNA would likely be relatively diluted and thus less likely to be detected but is an important consideration in the interpretation of results nonetheless.
The effect of time, however, did remain significant after controlling for environmental variation. It seems likely that this is largely due to interspecific differences in the timing of emergence and other life history traits of insects and other arthropods that are linked to seasonal events. No doubt the seasonal cues for such events go well beyond the simple set of weather signals we have analyzed. Periodic seasonal dynamics are clearly evident in other field studies of arthropod abundance and biodiversity (
Wolda 1978,
1988;
Stinson and Brown 1983;
Grøtan et al. 2012;
Hatosy et al. 2013;
Hallmann et al. 2017), but seasonal effects are often neglected in biodiversity studies.
Attaining a better understanding of the mechanisms that govern arthropod communities in agroecosystems has important management implications. If spatial variation in local plant composition proves to also have a strong influence on variation in the composition of arthropod communities, as our work suggests, then the focus of conservation efforts might be best placed on ensuring that a diverse set of plant communities are well represented across the landscape (
Economo 2011).
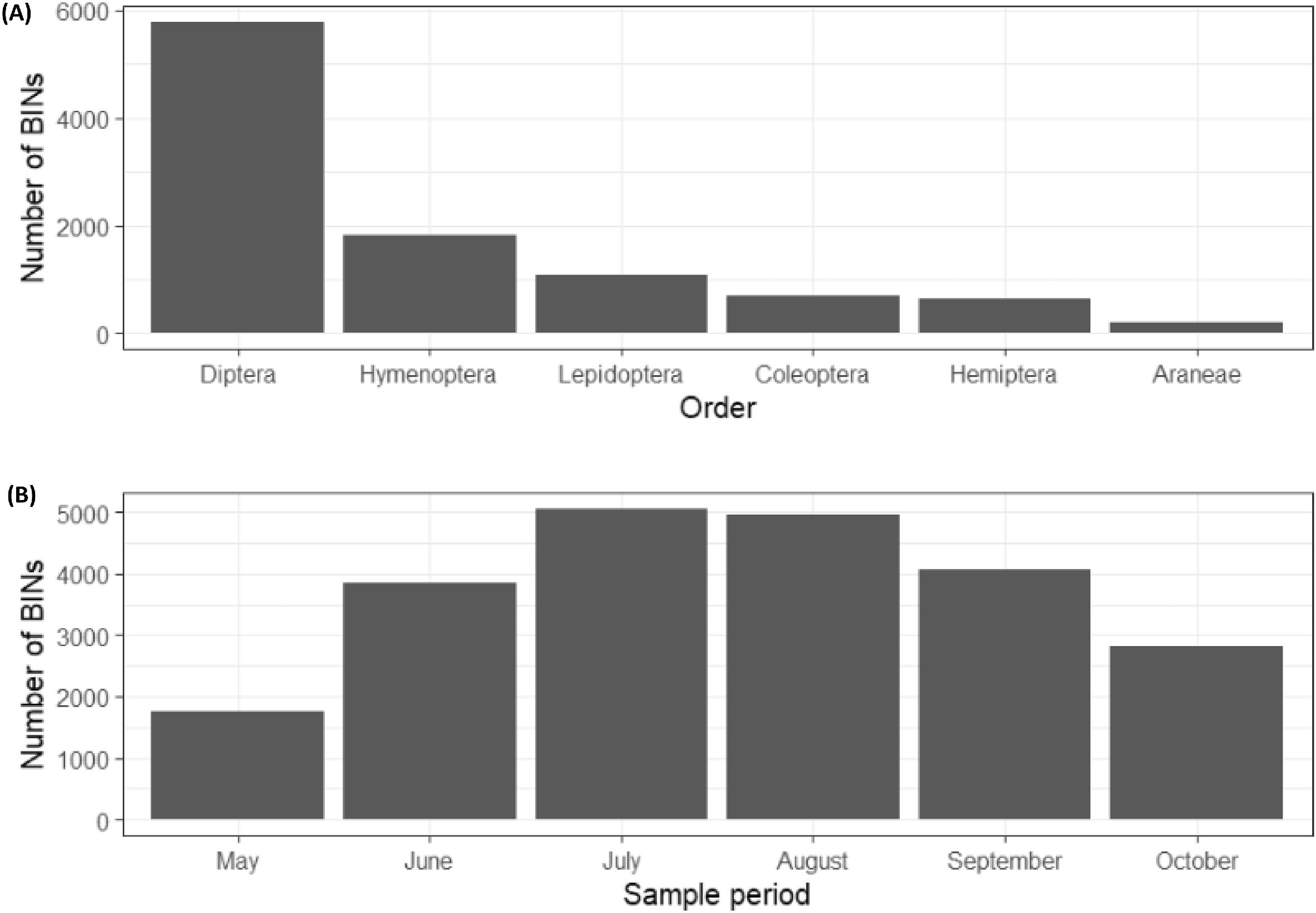
A significant achievement of our study was to provide a demonstration of how DNA metabarcoding can be used for large-scale terrestrial biomonitoring. During the course of our sampling, 3777 of the BINs sampled (26%) matched the DNA of species previously catalogued in the BOLD database. The fact that so many BINs of unknown species identity were recorded (74% of our sample) in a temperate North American region that has been widely sampled for biological diversity highlights the potential utility of bulk DNA metabarcoding as an exploratory lens to assess the limits of our taxanomic knowledge. Our ability to examine the composition of arthropod communities with such broad taxonomic coverage at this spatiotemporal scale was enhanced by the combined use of metabarcoding and Malaise traps (
deWaard et al. 2019;
Hausmann et al. 2019). Malaise traps are widely available, easily standardized, passive, and capture an enormous number of arthropods from a variety of taxa that represent many functional groups (
Karlsson et al. 2020). Metabarcoding provides a standardized method for species assignment even when a species has not been formally described, allows finer taxonomic resolution, speeds sample processing time, and is very cost-effective (
Cristescu 2014;
Bush et al. 2020). Using barcoding rather than morphological identification can also increase estimates of species richness and beta diversity by revealing cryptic species (
Brehm et al. 2016;
D'Souza and Hebert 2018), which allows for a more robust assessment to be made about the factors that drive variation in community composition (
Bush et al. 2020). The data generated from collaborative biomonitoring programs using a methodology such as this could create a vast repository of data that can address some of the most critical questions in community ecology facing us today (
Derocles et al. 2018;
Makiola et al. 2020). In particular, the advent of new techniques in network ecology can allow researchers to achieve a more mechanistic understanding of arthropod declines and their consequences for the rest of the ecosystem (
Petsopoulos et al. 2021).
There are a few important caveats to consider in interpreting the results of our study. First, our use of DNA metabarcoding could not capture some important information about these communities. Of particular relevance is the fact that DNA metabarcoding does not necessarily yield accurate measures of abundance (
Elbrecht and Leese 2015) and hence our arthropod data was analyzed as presence/absence. As well as this, functional information such as sex, life-stage, and body condition cannot be inferred by DNA metabarcoding (
Compson et al. 2020). For example, many species are only pests in one life stage and as such it would be valuable to know what life stage the incidence of the species refers to for the proper timing of pest management procedures throughout a growing season (
Crimmins et al. 2020). Recent work in this same system has demonstrated that deep learning from computer vision can be used to provide abundance and biomass estimates from photos of Malaise trap samples (
Schneider et al. 2021). Given the large proportion of singletons in our samples, DNA metabarcoding of Malaise trap samples would be an unacceptably crude way to identify candidate species of conservation concern. Future studies that link DNA metabarcoding of Malaise trap samples with such computer-vision based techniques to assess abundance at the species level could conceivably overcome some of these barriers.
PCR amplification is well known as a potential source of bias in metabarcoding studies and covered in detail elsewhere (see
Zinger et al. 2019). The primers chosen for this project have been demonstrated to amplify across a wide taxonomic range (most Arthropoda and many other invertebrate phyla), which covers the taxa most often collected by Malaise traps (particularly flying insects). Furthermore, any biases that are present are expected to be evenly distributed across samples and sites, and therefore should not affect the conclusions of this study.
Arthropods collected by Malaise traps must enter the head of the trap before falling down into a clear collection bottle, where they sit for one to several days before servicing and proper preservation in a freezer. This design does not readily collect airborne DNA and preserve airborne DNA well for subsequent detection. With that said, you might expect trace amounts of airborne DNA to enter the samples and possibly be detected through high-throughput sequencing. Much like other sources of environmental DNA (e.g., trace amounts of DNA carried on or inside specimens), the “noise” of airborne DNA is expected to be orders of magnitude lower than the “signal” of the target specimens in the sample; in most cases, this (and other) “noise” will be filtered out during data analysis.
It is also important to note that the placement of the traps (often within 100 of a second habitat type) could have resulted in edge effects, and that the local scale of the plant surveys may not have captured all possible relationships between explanatory variables. In particular, it is certainly plausible that at larger scales one might see stronger links between agricultural practices and arthropod communities. Future studies could address edge effects by a design that isolates habitat effects (e.g., placing all traps within a specific habitat rather than close to edge between habitats) and could address possible interactions between agricultural practices, plant communities, and arthropod communities in more detail by conducting plant surveys at entire-site scales. As well as this, our study sites were from a landscape characterized by a heterogeneous mixture of small farms, forest fragments, small wetlands, and grassland patches and our findings are therefore most representative of highly heterogenous agricultural settings. An important future research direction would be to further tease apart specific effects of different farming management practices on arthropod communities.