Introduction
The American lobster (
Homarus americanus) fishery is Canada’s most valuable fishery, generating $2 billion in commercial landings in 2021 in Atlantic Canada (
Department of Fisheries and Oceans 2022). This fishery generates thousands of jobs in rural areas and is significant for the economy and culture of Atlantic Canada (
FAO 2020). The present and future of the global crustacean fishery are influenced by several factors; however, disease is one of the greatest impediments to their long-term sustainability and growth. Shell diseases are the most common diseases that affect multiple crustacean genera globally. These diseases have been noted in crustaceans for at least 100 million years (
Klompmaker et al. 2016) and affect many, if not most, crustacean species. They present as lesions of the shell but could be caused by unrelated infections. These diseases can manifest themselves in wild populations, or in captivity, and can cause the death of these animals by secondary infections (
Cawthorn 2011).
Among
H. americanus, one of the most economically significant shell diseases is impoundment shell disease (ISD). ISD may occur in lobsters kept in live storage, especially during the winter (
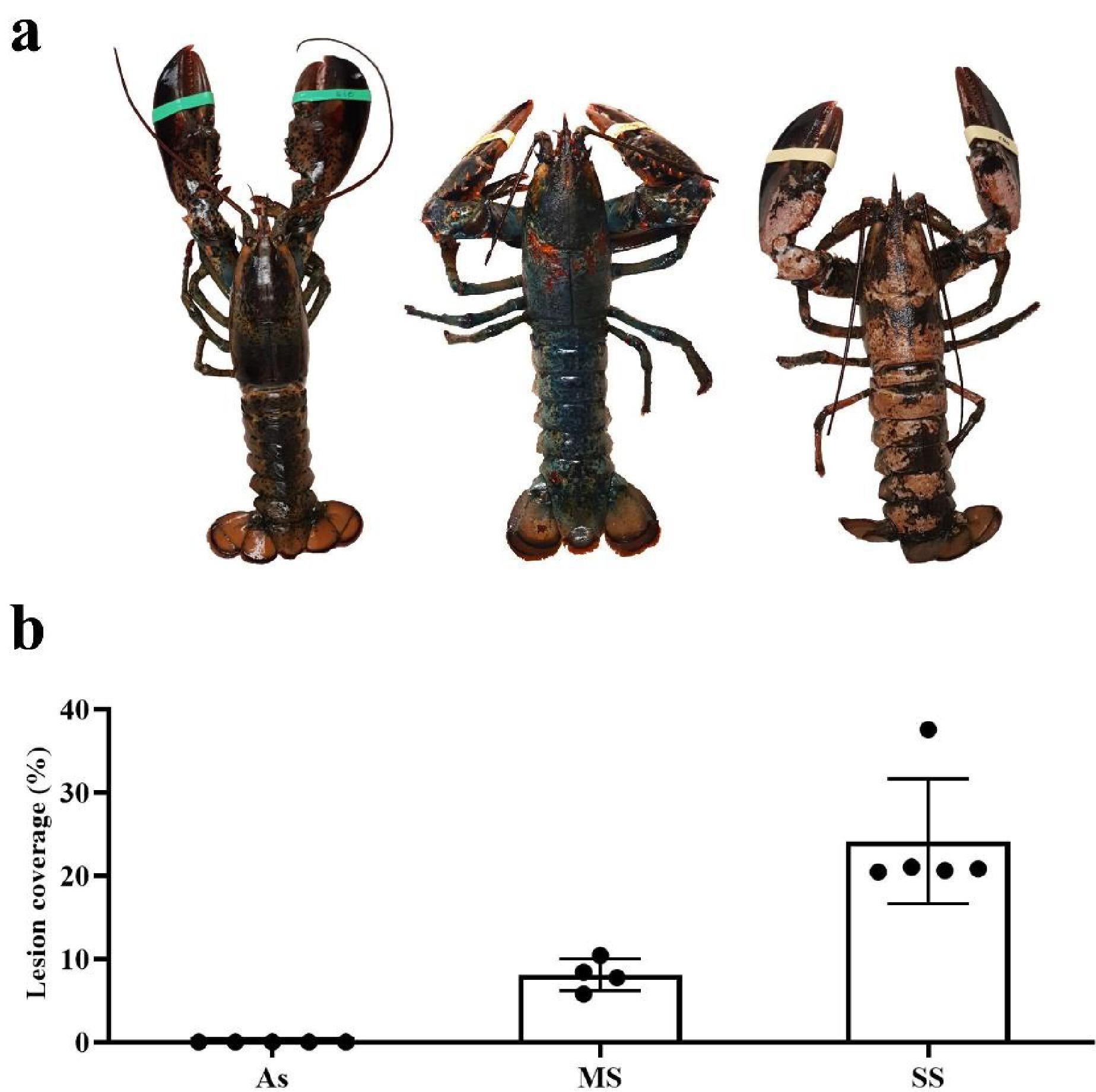
Smolowitz et al. 1992). The presence of ISD prevents animals from being sold through the live-market due to their unappealing appearance and, in more serious cases, can result in the mortality of the animal. It is characterized by the development of bilateral and symmetrical circular lesions around setal pores in the early stages of the disease, but lesions overlap and collapse to develop into a severe shell disease in advanced stages (
Fig. 1a) (
Theriault et al. 2008a). Although ISD was discovered in 1937, its causes were not fully characterized because many multifactorial physiological and environmental effects make identifying a single cause of shell diseases development difficult. Firstly, its development may not be due to a single microorganism, but rather a mix of effects caused by many microorganisms. Among the microorganisms found in ISD are mainly filamentous algae in early epicuticular erosions and, in advanced stages, a mix of fungi, bacteria, and protozoa (
Smolowitz et al. 1992). As with other shell diseases, chitinolytic bacteria are found at all stages of the disease (
Smolowitz et al. 1992). In addition to the cause of the disease not being a specific microorganism, this disease can be caused by opportunistic microorganisms from the natural microbiota of the shell when animals become more susceptible. This happens in the epizootic shell disease (ESD), a disease symptomatically similar to ISD that occurs in wild populations (
Meres et al. 2012).
Like all other crustaceans, American lobsters have an innate immune system to avoid pathogenic proliferation and infections (
Li and Wang 2021). The hepatopancreas is the most important immune organ in crustaceans, acting in crucial immune responses, such as phagocytosis by fixed phagocytes, and the production and release of systemic immune effectors (
Ridgway et al. 2006;
Sun et al. 2008;
Vogt 2019;
Yu et al. 2022). Little is known about the hepatopancreas immune responses during shell diseases in lobsters, where only one study evaluated the expression of hepatopancreas genes in lobsters with ESD (
Tarrant et al. 2010). The advancement of high-throughput transcriptomic studies in recent years has allowed an understanding of host–pathogen interactions in more detail, which is essential to understand immune responses against complex diseases such as ISD.
The aim of this study was to identify ISD-related immune molecules by comparing the hepatopancreatic transcriptome of asymptomatic (As), moderately symptomatic (MS), and severely symptomatic (SS) lobsters. Although these lobsters were subjected to the same impoundment conditions, they developed different degrees of disease or remained As, which is possibly related to their transcriptional profiles.
Discussion
This is the first study to use a high-throughput sequencing technique to understand
H. americanus responses to ISD. The objective of this work was to characterize the genes associated with the presence of ISD in lobsters exhibiting different degrees of ISD. This work was also the first to take advantage of the first version of the recently published genome of
H. americanus (
Polinski et al. 2021), aligning the reads to the genome instead of a de novo transcriptome and potentially revealing genes that would not be identified in this study without access to the genome. The hepatopancreas was chosen for this study because it is the most important immune organ in crustaceans, acting in crucial immune responses (
Ridgway et al. 2006;
Sun et al. 2008;
Vogt 2019;
Yu et al. 2022). Moreover, this organ is also responsible for glycogen storage (
Bonilla-Gómez et al. 2012), which is important because the animals from this study were stored for months and were unable to exhibit all of their natural behaviors. However, all animals had similar levels of glycogen reserves. Some studies showed that pathogens can be found in the haemolymph of lobsters infected with ISD (
Smolowitz et al. 1992). Once in the haemolymph, these microorganisms can spread to other organs, but the hepatopancreases of the animals in the current study were free from pathogens, showing that the disease was contained at some level at this stage.
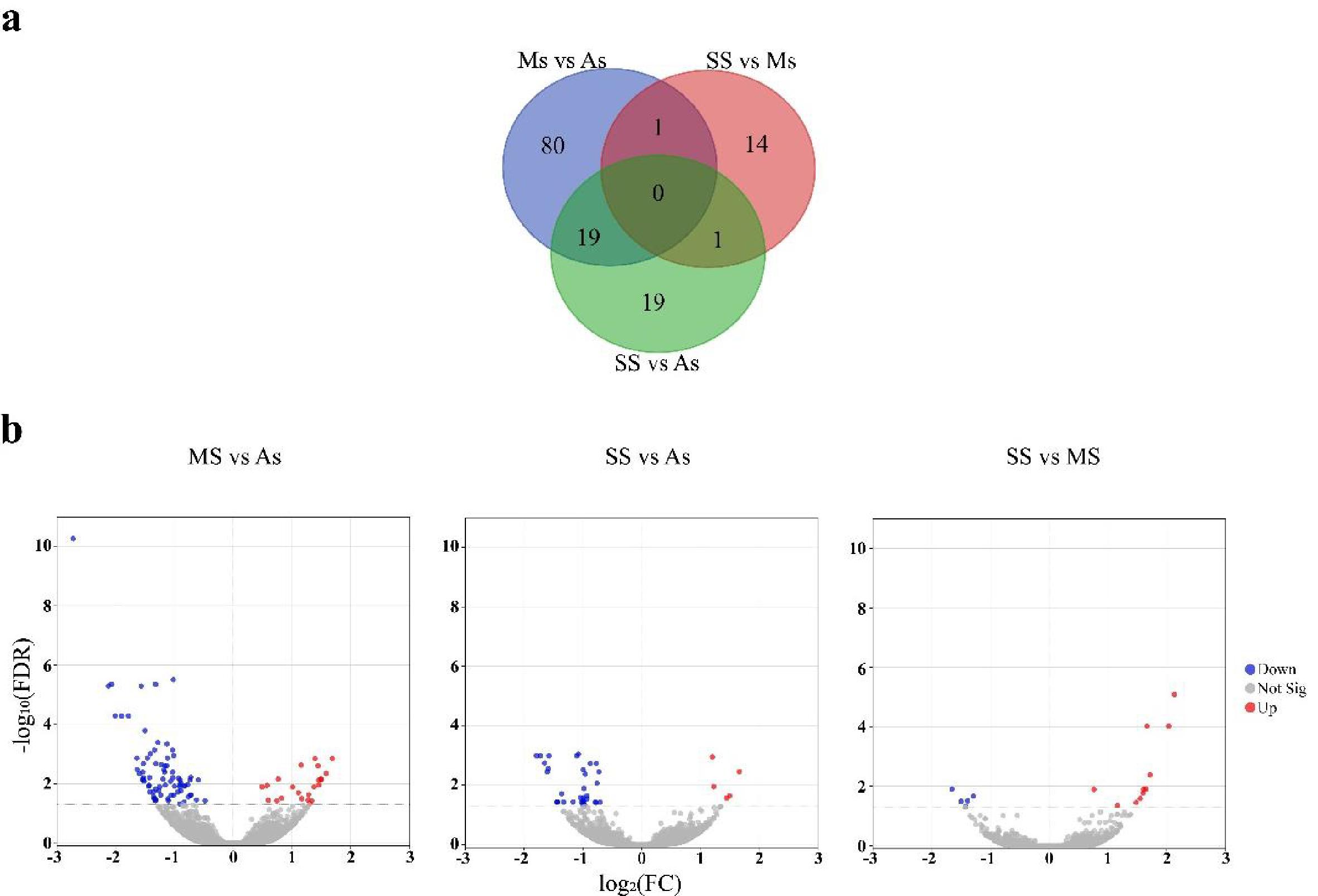
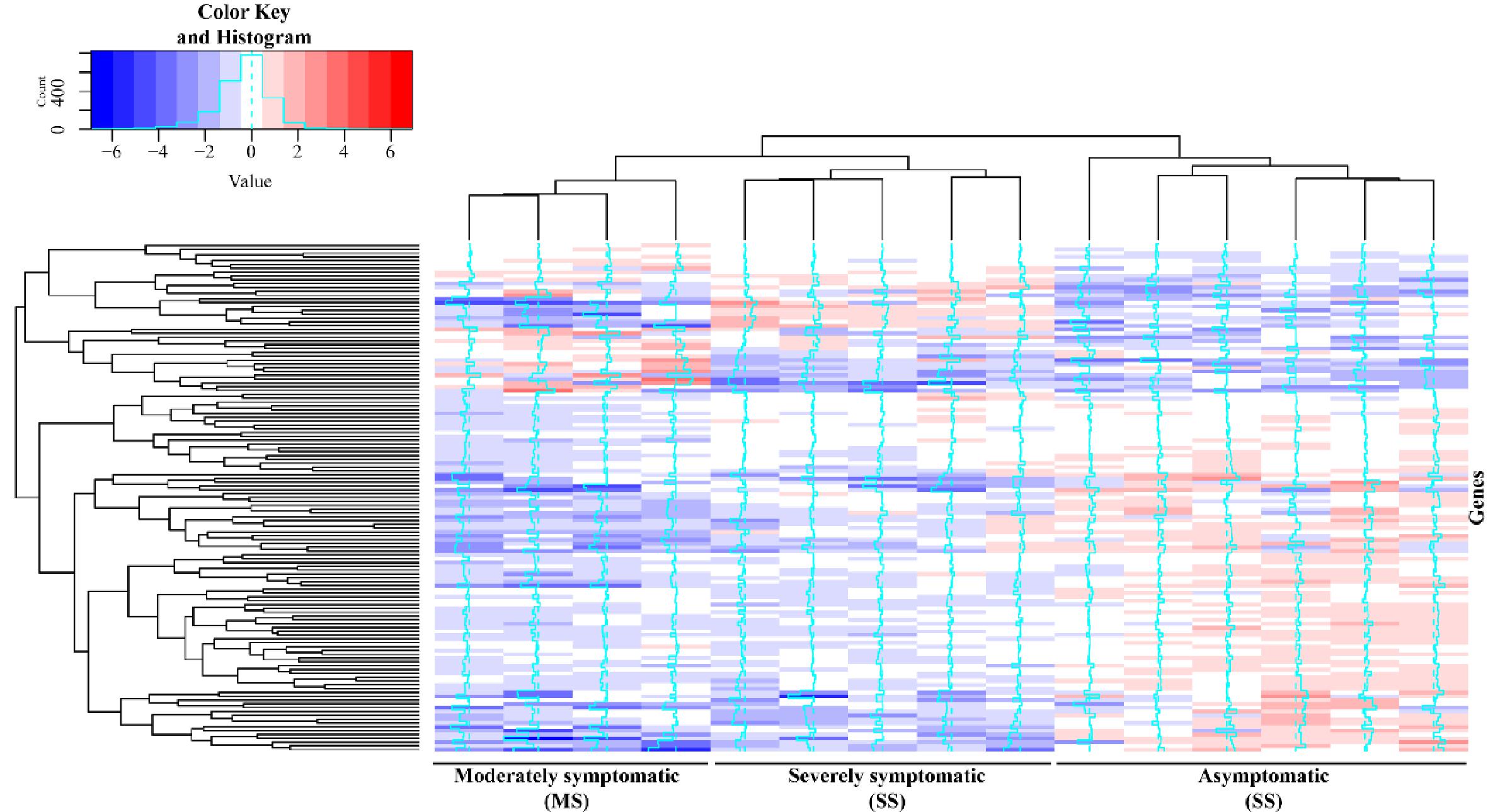
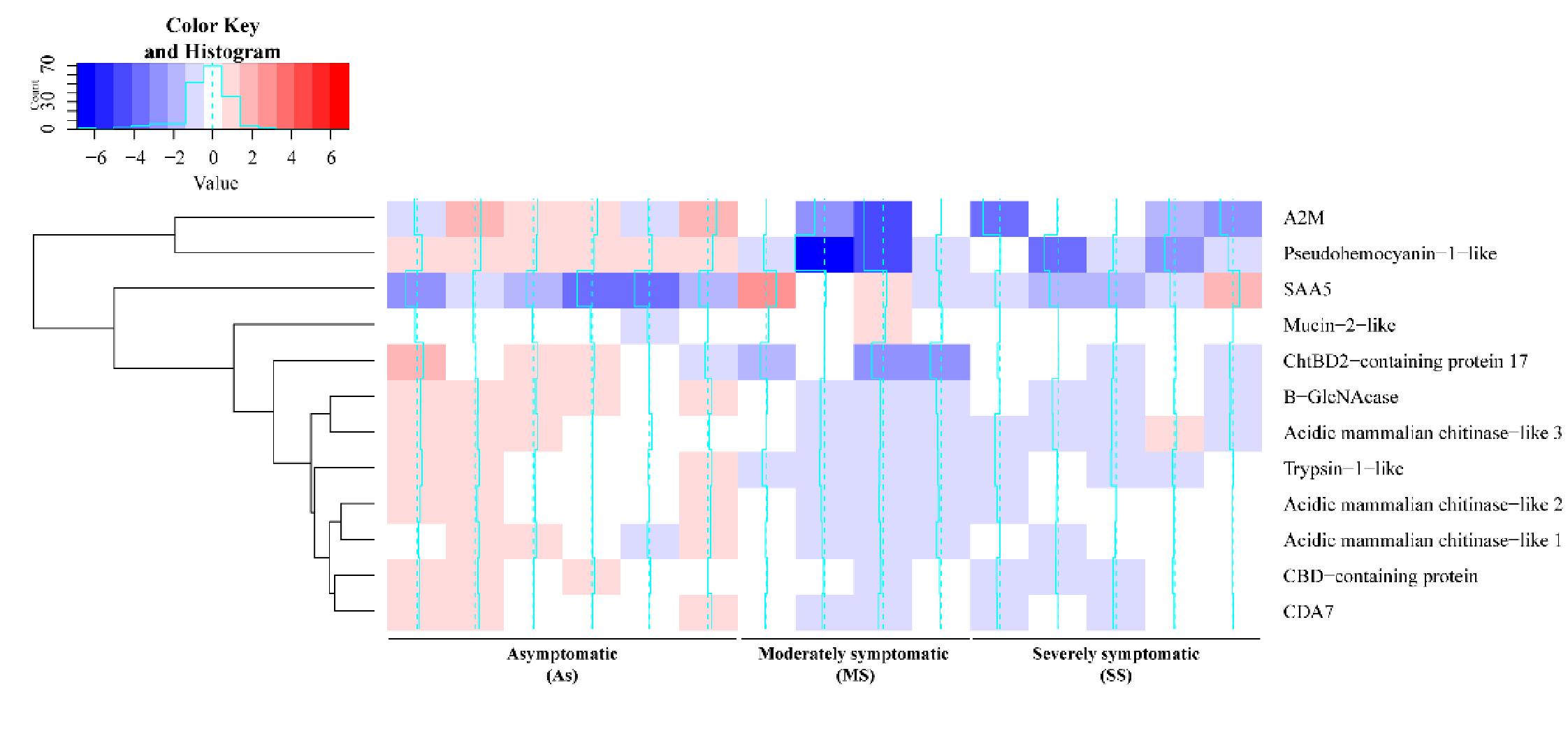
Most of the 134 DEGs potentially involved with ISD identified had higher transcriptional levels in the As group compared to the symptomatic groups (
Fig. 2). After an infection, microorganisms may evade the defense mechanisms of their host in different ways, such as controlling host gene regulation (
Cole and Nizet 2016). For example,
Dai et al. (2022) suggest that
Vibrio owensii hijacks the immune system of
Fenneropenaeus merguiensis by reducing the expression of C-type lectin and ficolin, resulting in reduced recognition and phagocytosis of the microorganism. In this sense, it is possible that the largest number of genes being less expressed in symptomatic animals in the present study is due to gene suppression caused by microorganisms associated with the ISD. Alternatively, it is possible that the difference in gene expression is the result of a greater ability of As animals to have a greater constitutive immune response against ISD. All animals in this study were subjected to the same biotic and abiotic factors, where some animals developed moderately to severe degrees of the disease, others remained As.
Tlusty et al. (2007) developed a hypothesis that it is the internal conditions of the lobster that make the animal resistant or susceptible to ESD, although several environmental and pathological factors also contribute to its development. Therefore, the transcriptional profiles observed in the present study could be part of the internal conditions of these animals contributing to the resistance or susceptibility to ISD development. Transcriptional profiles were previously associated with resistance and susceptibility against diseases in shrimp (
de Lorgeril et al. 2005). In line with this idea,
Theriault et al. (2008b) examined physiological parameters of
H. americanus prior the storage in pounds and evaluated the development of ISD for 120 days. Interestingly, lobsters with lower levels of total protein developed more ISD compared to lobsters with higher levels of total protein (
Theriault et al. 2008b).
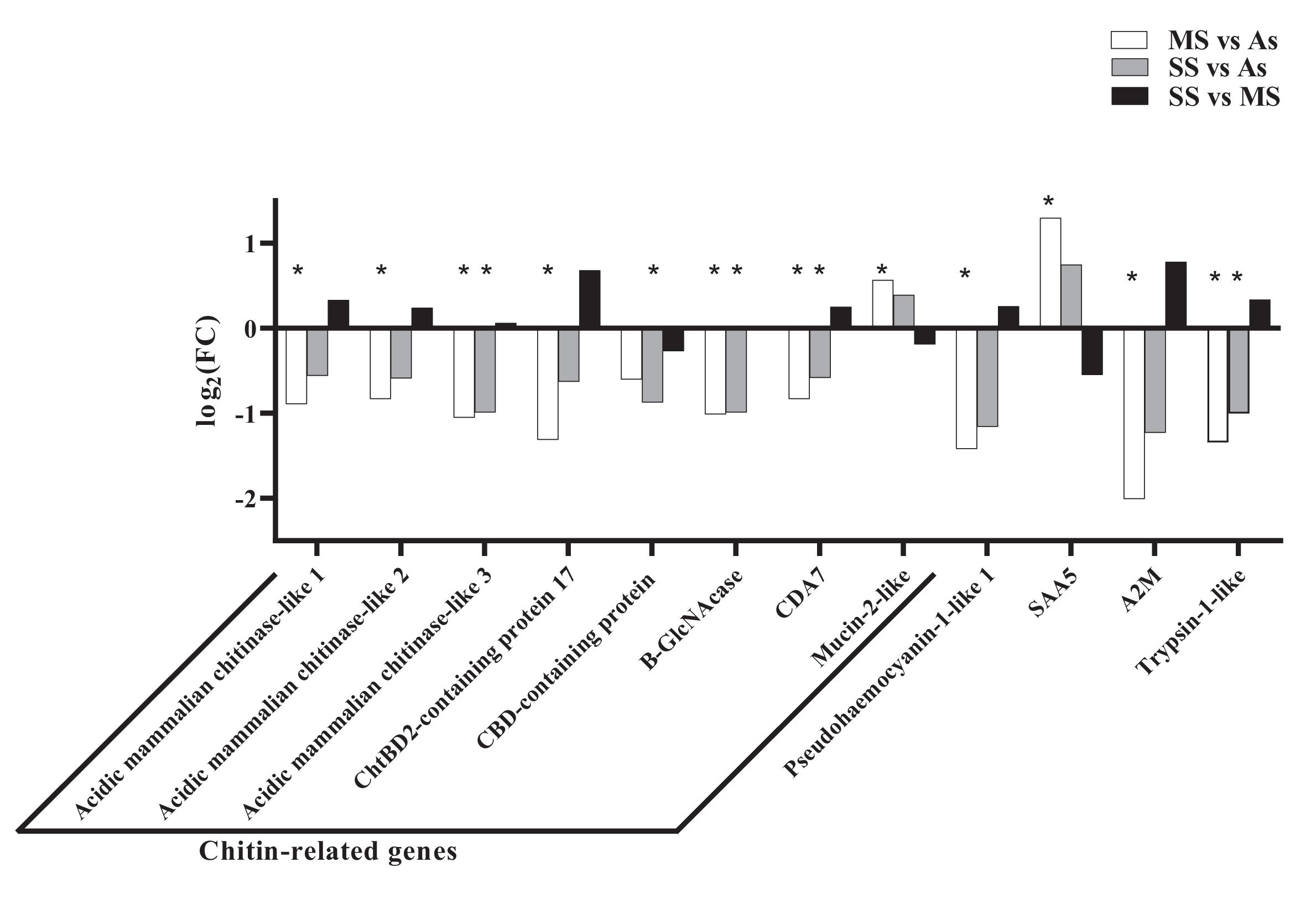
Among the DEGs found in this study are several chitin-related genes, such as three chitinases and one chitooligosaccharidolytic β-N-acetylglucosaminidase-like (B-GlcNacase). While chitinases degrade the chitin molecule into chitin-oligosaccharides (GlcNAc)
n, B-GlcNacase is responsible for cleaving these oligosaccharides into monomers of N-acetylglucosamine (
Merzendorfer and Zimoch 2003;
Yang et al. 2008;
Qiu et al. 2022). These enzymes are distributed in different organisms, since chitin is the second most present biopolymer in the world, participating in different physiological processes (
Qiu et al. 2022). Chitinases have three primary functions in crustaceans: participating in the molting and growth processes of these animals, helping in the digestion of foods that contain chitin, and directly or indirectly participating in defense responses against bacteria, fungi, and viruses (
Zhang et al. 2021). Although these molecules have the ability to kill some chitin-containing pathogens through their catalytic activity (
Zarei et al. 2011;
Seo et al. 2016), they seem to play a major role in direct and indirect immune regulation (
Niu et al. 2018;
Zhang et al. 2021). The
LvChi5 chitinase from
Penaeus vannamei does not show in vitro activity against
Vibrio parahaemolyticus or
Escherichia coli, but expression of
LvChi5 was induced against
V. parahaemolyticus, white spot syndrome virus (WSSV), lipopolysaccharides (LPS), and poly(I:C) (
Niu et al. 2018).The absence of
LvChi5 through silencing by RNAi caused an increase in the mortality of
P. vannamei against infections with WSSV and
V. parahaemolyticus. Moreover, silencing of this gene also resulted in transcriptional inhibition of immune genes such as antimicrobial peptides, transcriptional factors, and other proteins with antibacterial or antiviral activity (
Niu et al. 2018). Chitinase transcriptional levels of WSSV-resistant
Penaeus japonicus were higher than virus-sensitive animals (
Pan et al. 2005). The three chitinases differently expressed found in the present study are encoded by different genes and share an aminoacidic identity ranging from 54.2% to 68.2% to each other. Chitinases are very diverse molecules, being divided into two families with multiple groups according to sequence homology and catalytic mechanism with multiple functions, which could be pathogen-specific or gene-specific regulations (
Zhang et al. 2021). Therefore, the activity of some chitinases may be mainly due to immune regulation and these genes could play a major role in the ISD development through multiple routes.
As well as chitinases, B-GlcNacases comprise a diverse family of intracellular, secreted, or transmembrane molecules that appears to participate in immune responses (
Zhang et al. 2021;
Soto-Rodriguez et al. 2022). Gene expression of a GlcNacase from
Exopalaemon carinicauda increased in the hepatopancreas after infection with
V. parahaemolyticus and
Aeromonas hydrophila (
Sun et al. 2018). In mammals, GlcNacase is one of the preformed mediator lysosomal enzymes, which is stored in some granulocytes and is released upon infection (
Prabhuling et al. 2021). These molecules are also associated with intercellular communication during immune responses in mammals (
Prabhuling et al. 2021). In cell cultures of
Drosophila melanogaster, silencing of a lysosomal GlcNacase by RNAi resulted in an increase of mycobacteria growth (
Koo et al. 2008). In
Bombyx mori, GlcNacase activity is increased during haemocytic degranulation after parasitism, indicating the release of this molecule to fight the parasite directly or indirectly (
Prabhuling et al. 2021). The mucin-like protein is found in the membrane of the hepatopancreas and is associated with protection against bacteria (
Soto-Rodriguez et al. 2022), but some contradictory studies suggest an importance of mucin-like protein for bacterial establishment (
De los Santos et al. 2022). It would be important to understand the role of mucin-2-like protein during ISD in future studies, since this was the only chitin-related gene that was more expressed in symptomatic animals.
A2M protein is a class of protease inhibitors abundant in the haemolymph of invertebrates, and acts as a multifunctional protein (
Chaikeeratisak et al. 2012). They act as protease inhibitors neutralizing pathogenic proteases important for bacterial establishment (
Armstrong 2010). However, these molecules are also involved in other immune responses, such as phagocytosis, prophenoloxidase activating systems, and the haemolymph clotting system (
Chaikeeratisak et al. 2012). The gene modulation of A2M in the hepatopancreas and haemocytes of crustaceans during bacterial and viral infections has been reported (
Pongsomboon et al. 2011;
Ren et al. 2019). Previous studies found that the A2M expression was higher in symptomatic animals with ESD (
Tarrant et al. 2010), which is contrary to the findings of this study (
Tarrant et al. 2010). This suggests that, although ISD and ESD are similar in appearance, some immune responses against these two infections may be different.
Trypsin-1-like was also more expressed in As animals compared to others, which may be indicative of an immune response against ISD pathogens. Trypsin is related to metabolic processes but may also contribute to defense mechanisms in crustaceans in the form of a serine protease. Interestingly, the gene expression of trypsin-1 is frequently upregulated during infections in crustaceans (
Clark et al. 2013a,
2013b,
2013c;
Li et al. 2018;
Yu et al. 2022). Trypsin may also be associated with the cleavage of haemocyanin into haemocyanin-derived peptides with immune activity. The regulation of trypsin against infections is related to the generation of haemocyanin-derived peptides in
P. vannamei and its silencing by RNAi reduces the amount of haemocyanin-derived peptide. In addition, in vitro experiments have shown that recombinant trypsin hydrolyzes haemocyanin and the generated fragments have bacterial agglutinating capacity (
Li et al. 2018). In line with this idea, a pseudohaemocyanin gene was also more expressed in As animals compared to symptomatic animals in this study. Pseudohaemocyanin function is not well understood, and there is no in vitro study showing its cleavage to produce fragments with antimicrobial activity. However, there is evidence of this molecule being cleaved, and the possibility of pseudohaemocyanin-derived peptides with antimicrobial activity should not be discarded (
Terwilliger et al. 2005). Therefore, it is possible that the increase in trypsin and pseudohaemocyanin results in an increase in the number of fragments with antimicrobial activity to prevent the establishment of the disease.
Acute phase serum amyloid A or SAA is a protein present in the haemolymph of crustaceans associated with the acute phase of infections and is used as a biomarker in the medical and veterinary fields (
Cray et al. 2009). In this work, the SAA gene was more expressed in MS animals compared to the As group (FDR < 0.05), and this trend was also observed without statistical difference in the comparison between SS and As animals (FDR = 0.7). These results suggest that MS animals are at the acute stage of the disease, whereas SAA levels in SS animals have started to decline. This could also indicate that As animals are, in fact, in a healthy condition. Although SAA is classically a biomarker of the acute stage of a disease, this gene is also upregulated in moribund lobsters infected with the Gram-positive bacteria
Aerococcus viridans var.
homari and the ciliated protist
Anophryoides haemophila (
Clark et al. 2013a,
2013b). Altogether, it is possible to suggest that, despite having an advanced stage of ISD, the acute phase of the disease is over in SS animals.