Introduction
Over the past 5 years, there has been a significant investment in developing digital research infrastructure, data management support, and establishing data sharing best practices to promote accessibility and usability of research data in Canada. For example, Canada’s Tri-agency, which is composed of the Canadian Institutes of Health Research (CIHR), the Natural Sciences and Engineering Research Council, and Social Sciences and Humanities Research Council, released a data management policy (
Government of Canada 2021), which requires prospective applicants to submit a data management plan and indicate where they will deposit their data. In addition, the Digital Research Alliance of Canada (Alliance) was formed to support digital research infrastructure including data repositories as well as research data management (RDM) software and support for Canadian researchers. This organization has established and supports national data sharing platforms, including the Federated Research Data Repository, Lunaris, a national Dataverse repository, and a discovery service to index, store, and provide access to Canadian research data from across disciplines and sectors (
Digital Research Alliance of Canada 2022b,
2022c). Finally, the federal government released a Roadmap for Open Science in 2020, calling for all government-funded research outputs to be made openly available by default (
Government of Canada 2020a).
Finding and accessing data that is restricted or sensitive remains a common challenge for researchers despite these widespread efforts to improve data sharing. This challenge is enhanced by the fact that restricted data are by their very nature more difficult to access. For the purposes of this study, we define restricted data as data that are not immediately accessible because they are restricted due to commercial, ethical, or legal reasons, or they are only available upon request.
Recent studies have shown that author-reporting on the availability and accessibility of restricted data in publications is poor, particularly with respect to providing clear language and information in journal data availability statements (
Read et al. 2021;
Gabelica et al. 2022;
Page et al. 2022). A previous study completed by the corresponding author found that when CIHR-funded authors reported that data could be shared or an application was required, none provided details about how to access the data (
Read et al. 2021).
Gabelica et al. (2022) encountered similar issues when trying to access data from authors who stated in their data availability statements that their data were available upon request; only 6.8% of authors provided the requested data.
Another set of challenges arise from poor infrastructure for searching for, locating, and requesting restricted access data, specifically poor search interfaces (or lack thereof), difficult-to-use software, a lack of standardization for how to submit data requests, and a lack of support from restricted data stewards (
Nancarrow 2013;
Choudhury et al. 2014;
Sarwate et al. 2014;
van Schaik et al. 2014;
Lugg-Widger et al. 2018;
Rahimzadeh et al. 2018;
Garrison et al. 2019;
Bonomi et al. 2020). More generally, researchers have reported difficulties in both understanding and navigating the data application process (
Sydes et al. 2015;
Siu et al. 2016;
Ho et al. 2018;
Prince et al. 2018;
Rahimzadeh et al. 2018;
Bekemeier et al. 2019;
Saulnier et al. 2019;
Knosp et al. 2022). For the purposes of this study, we define data stewards as individuals or organizations responsible for dataset documentation, quality, storage, preservation, and access in any given data source; the responsibilities of a data steward may also include managing the technical infrastructure used to house those datasets (
CODATA no date).
These barriers to discovering, accessing, and using restricted data have resulted in several consequences: researchers have reported that they may limit their research questions to data that can be more easily accessed (
van Schaik et al. 2014); researchers may invest a substantial amount of resources (e.g., time and money) into acquiring data that cannot be easily found and/or used (
Siminski et al. 2021); and researchers have highlighted that academic and non-academic research (particularly in the health sector) is strongly limited when restricted data lack sufficient means to facilitate discovery and access (
Lugg-Widger et al. 2018;
Rahimzadeh et al. 2018;
Pongiglione et al. 2021).
While many of the studies above have examined the high-level ethical and legal barriers to restricted data sharing, few (
Leahey 2014) have explored the barriers that data providers across sectors face when trying to make their datasets discoverable and accessible. To help address these barriers, a Canadian national working group was formed to identify and evaluate the challenges related to discovering and accessing restricted data in Canada and to inform the ongoing development of Canadian infrastructure for research data. To that end, this study was undertaken to answer three primary research questions:
RQ1: What types of Canadian access-limited data sources include datasets that could be used for research purposes?
RQ2: Based on a sample of Canadian restricted health data sources identified in RQ1, how well do these sources make their data discoverable and accessible?
RQ3: What are the challenges associated with discovering and accessing restricted data from the sample of Canadian health data sources identified in RQ1?
The answers to these questions provide an opportunity to address barriers associated with finding, accessing, and using restricted data while examining it through a national lens. Ultimately, the findings identified in this study can inform how Canadian restricted data sources and the Canadian RDM landscape can work toward improving the discovery, access, and use of these data. While this study focuses on the Canadian RDM landscape and restricted data sources, it can be used to inform progress toward improving the discovery, access, and use of both Canadian and international data.
Results
Canadian landscape of access-limited data sources
Our scoping exercise was successful in identifying 137 access-limited data sources. Of these data sources, 49.6% (n = 68) were provincial in scope, 48.9% (n = 67) were classified as national, 2.0% (n = 3) were from the territories, and 1.5% (n = 2) were classified as international with Canadian datasets hosted within them. Each data source may have had more than one geographic category assigned to it.
With respect to the sectors these data sources represent, 61.3% (n = 84) were from the government, 26.3% (n = 36) originated from academic institutions, 15.3% (n = 21) were from the non-profit sector, and 4.4% (n = 6) were hosted in the private sector. 10.9% (n = 15) of data sources were classified as “other”, as they did not fit within the other predefined categories. Categories assigned to each data source were not mutually exclusive.
In terms of the disciplinary focus of the data sources identified, 40.8% (n = 56) were categorized as being related to medical, health, and life sciences, 16.8% (n = 23) were considered general purpose, 16.8% (n = 23) were from the natural sciences, 5.8% (n = 8) were from the social sciences, 2.2% (n = 3) were from agricultural and veterinary sciences, and 2.2% (n = 3) were from engineering and technology. The 16.8% (n = 23) of data sources that were labelled as general purpose represented data provided by provincial government ministries and agencies and included a variety of categories within our predefined list, including but not limited to government administrative records, the census, and parliamentary debate transcripts. 15.3% (n = 21) were classified as other as their focus was outside the scope of the predefined categories used in our analysis. For example, many data sources drawn from Canada’s National Research Council focused on property management, purchasing and requisition files, and business opportunity data that did not fit within either of the general purpose or discipline-specific categories. The complete list of data sources identified is available in Supplemental File S4.
Health data source scoring results
From the health data sources identified, 48 of the 55 data sources were included in the final analysis. Seven data sources were excluded because they either only accepted requests from individuals for their own personal health record data (n = 4) or the data source became inaccessible during our study due to broken URL links (n = 3).
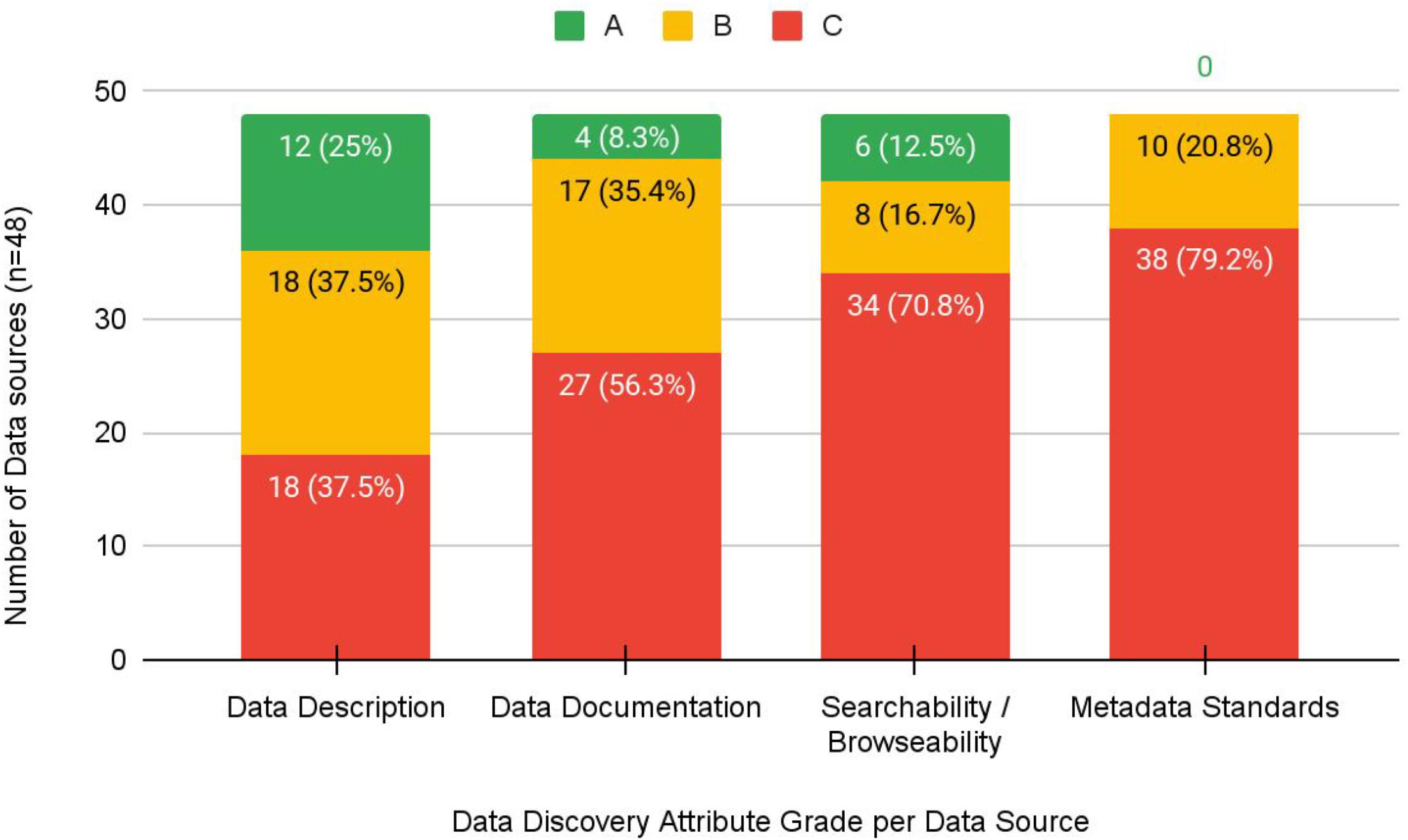
The grading exercise identified that 42% (
n = 20) of data sources did not receive an “A” grade in any category; however, 44% (
n = 21) received an “A” grade in two or more categories. The majority of datasets received a “C” grade for a lack of metadata standards (38/48, 79%), an inability to explore and discover datasets through searching and browsing (34/48, 71%), or lack of data documentation to support interpretability and reuse (27/48, 56%) (
Fig. 1). Descriptions of datasets themselves fared better in that 25% (
n = 12) received an “A” grade, and 37.5% (
n = 18) received a “B” grade.
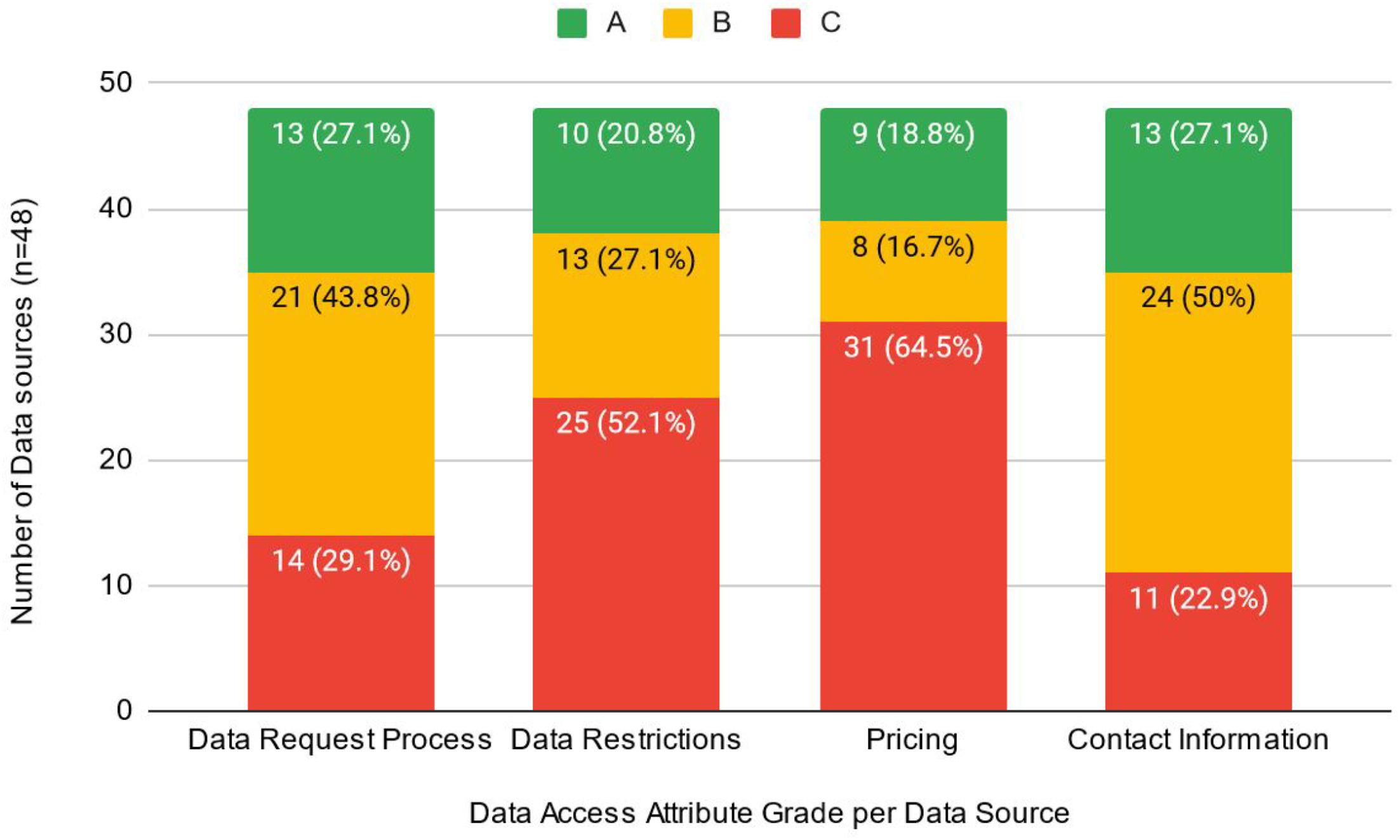
The absence of or lack of clarity with respect to pricing information (31/48, 65%) and vague or non-existent information related to dataset restrictions (25/48, 52%) were identified as key barriers to the data access request process (i.e., received a “C” grade). It is likely that many of the 31 sources without information about pricing are accessible for research purposes at no cost; however, providing this information to potential users is an important part in streamlining access. The actual description of the data request process, however, identified that 70.8% of data sources received an “A” (13/48, 27%) or “B” (21/48, 43.8%) grade in this category. Similarly, contact information was generally well described, as only 22.9% (
n = 11) received a “C” grade (
Fig. 2).
The degree to which individual data sources were consistent across discovery and access attributes was considered “fair” according to Kendall’s tau ranked test (tau-b = 0.31, p = 0.0059), indicating that data sources with higher discovery grades also had higher access grades on average relative to the other data sources in our sample.
Discussion and conclusions
This study provides preliminary insight into the discovery and access characteristics of restricted health data sources in Canada. In particular, this study highlights three key barriers associated with data of this kind that both mirror findings in previous research and introduce new complexities to the restricted data sharing landscape. First, this study calls attention to the challenges with discovery and accessing data due to a lack of sufficient infrastructure. Second, not a single data source received an “A” grade for metadata; for the majority of data sources, metadata were either sparse or non-existent. Finally, the availability of documentation for facilitating the reuse of datasets was lacking, resulting in an inability to understand or interpret data in the sources we identified.
To forge a path toward removing barriers in these three areas, we elaborate below on the challenges associated with these areas and suggest how to improve and support the discovery and access of restricted data in relation to them. In addition, we suggest approaches for examining data sources beyond the health sector to provide a more holistic view of the Canadian restricted data sharing landscape.
Improving restricted data infrastructure for discovery and access
Infrastructure was observed across the sources at varying levels ranging from individual research projects, institutional websites, large scale research organizations with support for data access, regional health data centres, and government data access programs. This infrastructure diversity reflects variations across research disciplines, institutions, and jurisdictions that may govern restricted data sharing, and highlights the difficulties of standardizing discovery and access to these data. The limited availability and high variability in workflows across data sources can lead to challenges around standardization, equitable access, and sustainability at the human, project, and technological levels. Without consistency and standardization across restricted data sources, datasets will remain difficult to find, access, and use. In a worst-case scenario, data sources that are under-resourced may result in data loss—an issue we encountered during our study as three sources became inaccessible during our analysis.
This study highlights that data sharing infrastructure in Canada does not adequately support making restricted data FAIR (findable, accessible, interoperable, reusable) (
Wilkinson et al. 2016). The ideal state of restricted data discovery and access infrastructure has not been fully defined or envisioned yet and requires a variety of stakeholders and experts to come together to define relationships and roles across institutions and jurisdictions. Researchers need access to reliable, secure, institutionally approved infrastructure and workflows for storing, sharing, and preserving research data to comply with funder and/or journal policies, and to facilitate reproducibility and reuse of restricted data across sectors. Without sufficient infrastructure, these data will remain hidden, difficult to access, and may even be lost due to lack of support. For an infrastructure model that supports restricted data discovery and access to be successful on a national scale, we recommend that government (specifically provincial/territorial health bodies), academic institutions, and national data management initiatives (e.g., the Alliance) work together to develop a standardized model that supports restricted data discovery and establishes standard workflows for accessing restricted data for research purposes.
Developing and adopting robust discovery and access metadata standards
We found no consistent metadata describing restricted datasets or their access-specific requirements among data sources we assessed. While some sources included non-standardized metadata elements specific to restricted data, the use of specific metadata schemas was absent. Our grading results demonstrate that data sources provide reasonably good description of their data and the request process to access them; however, a lack of metadata with which to structure or disseminate this information impedes their ability to be discovered by the research community.
Restricted data sources could adopt existing metadata schemas to positively impact discovery, use, and standardization. The inclusion of structured metadata would help researchers by reducing time and resources expended on identifying valuable datasets. The findings from our grading exercise emphasize this, as over half of the data sources did not provide any information about who is eligible to access their datasets. Structured metadata would also improve discovery by increasing the potential for restricted data to be harnessed by national aggregators. National aggregators play an important role in making data discoverable, accessible, and therefore, reused; however, the extent to which they can perform this function is limited by the lack of standardized, openly available metadata. Those interested in using restricted data can use metadata harvested by aggregators to filter and sort data sources and datasets, so that they can identify data that suit their needs.
Presently, existing metadata standards may not be sufficient to adequately describe restricted data, specifically with respect to the data access request process. Although generalist metadata schemas include generic elements to describe access restrictions, guidance for their application is often left open-ended; for example, Dublin Core’s “accessRights” element does not provide sufficient structure to describe access procedures (
Dublin Core Metadata Innovation 2020). In practice, the values of these elements are often free text and do not have consistent values. Moreover, vocabularies to describe access—such as the
Confederation for Open Access Repositories Access Rights vocabulary (2021)—describe the access status of a resource (such as “open access” or “restricted access”) but do not describe the
conditions of access restrictions (such as who can access the resource or at what cost).
Data sources would benefit from standard metadata elements to describe data access procedures, as expenditures of resources deployed to make data discoverable, to create data governance and access frameworks, and to ensure compliance may be reduced by addressing current ambiguities in best practices. Researchers would also benefit from access procedures being clearly defined in standardized metadata. For example, the cost to access restricted data varies greatly among data sources. If cost were to be included as a metadata element by multiple data sources, a researcher could search within a national aggregator to identify datasets that may be accessed at no cost. This would greatly improve the ability of researchers to identify data appropriate for their particular study in relation to its funding.
One way for restricted data sources to adopt existing metadata schemas and improve discoverability is to connect with global research infrastructure. Digital object identifiers (DOIs) are a type of persistent identifier commonly used for research outputs—such as data and publications—with accompanying metadata. If restricted data sources were to begin registering DOIs for their datasets, they would be providing standardized metadata—for example, according to the
DataCite Metadata Schema (2021)—and this metadata would be publicly searchable and usable by aggregators.
Data are valuable insofar as researchers are aware they exist and know they may access them. The gaps in existing metadata practices we found among data sources show the importance of how information about data sources and data access is communicated, not merely what is communicated. Existing metadata schemas are necessary but need to improve their ability to improve discovery of restricted data; they currently do not provide sufficient structured metadata to document restricted data access procedures.
Based on the lack of metadata identified in the 137 data sources from this study, we believe a logical first step to improving their discoverability would be to have them develop metadata that align with a schema used by national aggregators like Lunaris (
Digital Research Alliance of Canada 2023)—Canada’s research data discovery platform—so that they can be harvested by those systems. Creating metadata for these data sources would in turn make them harvestable in other systems such as Google Dataset Search.
Another recommendation resulting from this study is that existing metadata standards bodies like DataCite (
DataCite 2021), the
Data Documentation Initiative (2023), and the W3C Data Catalog Vocabulary (
World Wide Web Consortium 2023) must begin to develop more robust metadata to account for the restricted data access request process. Without metadata to sufficiently describe that process, it may be findable, but the access challenges researchers face determining whether they are eligible to access a dataset will persist.
Support and training for data documentation best practices
From our grading exercise, only four data sources received an “A” grade for the “Data Documentation” attribute. This component of discoverability is critical for researchers as, without the ability to view and/or interact with the data, a researcher cannot know whether the data are suitable to answer the research question unless they are sufficiently well documented.
Careful and deliberate documentation of data can be onerous and time consuming but provides the minimum benefit for the researcher/organization who collected the data and a maximum benefit for secondary users of the data. These incentives are clearly reflected in the sub-optimal documentation of the health data sources we investigated. The organizations that fared well in our grading exercise were evidence of this as large organizations with many administrative staff were shown to have well-documented data, compared to data sources from small research groups run mostly by academics who had little to no documentation.
While this study has focused primarily on the issues of discovery and access of restricted data, the lack of documentation found in the health data sources indicate that much of the data we identified may not be usable. Better training and support for data stewards to develop strong data documentation are needed to facilitate secondary use of restricted data, including but not limited to user guides, data dictionaries, documented code, and readme files. Funding bodies implementing data management or sharing requirements, national organizations like the Alliance who promote data management best practices, and academic institutions who host or support restricted data sources should invest in the development of training programs for the data stewards responsible for these sources to ensure that they have the skills necessary to develop sufficient documentation to enable data repurposing or reuse.
Exploring the national landscape of access-limited data sources
Returning to the first step of this study, our preliminary scoping of access-limited data in Canada identified data sources from across sectors and disciplinary areas, capturing a diverse landscape of data related to the sciences, infrastructure, environment, demography, agriculture, and others. While our analysis was restricted to health-related data sources within our sample, we see value in applying (or adapting) our methodological lens, materials, and findings to an exploration of the remaining data sources that were not our focus. Questions worth investigating include the following: Do discovery and access trends among restricted data sources vary widely by field/topic and sector? How well do non-health sources perform against our grading rubric? Is a standardized framework for making disparate restricted data more discoverable and accessible a desirable or possible outcome? At minimum, a thorough comparison of the discovery and access characteristics of non-health data sources in our inventory will provide insight into the challenges users and curators face when interacting with data that cannot be shared openly.
This study provides a preliminary evaluation of Canadian restricted health data sources’ ability to make their data discoverable and accessible and highlights key gaps that need to be addressed by the RDM community to improve their use. As the Canadian government continues to release policies that call for data to be FAIR, data sharing infrastructure must accommodate the complexity of needs required for restricted data to be found, acquired, and used. Without suitable digital infrastructure, better metadata, standardized workflows for accessing data, and robust documentation to facilitate secondary use, these valuable datasets will remain hidden, insufficiently supported, and underutilized.
Limitations
We acknowledge that the 137 data sources identified in our study do not represent all Canadian access-limited data sources, and some data sources may have been omitted. That said, our team of experts in data discovery and access searched as comprehensively as possible and reached out to experts in the field to identify sources we may have missed. One area that may be less represented than others is in the province of Quebec, where we relied on the expertise of others to provide us with French data sources.
With respect to the grading exercise, our scores were limited to what was available from the public version of the data source only. It may be possible that additional description, metadata, and documentation are available once a data request is made, and data are acquired. We did not want to burden data sources with false requests, which is why we omitted this step. Furthermore, our approach was meant to mimic someone who was interested in the data and browsing for it in the data source as if they were looking for data for the first time.