Introduction
Global plastic production between 2016 and 2020 exceeded 300 million tonnes annually (
Plastics Europe 2021). In Canada, plastic production is a large industry that contributes $35 billion to the national economy and employs over 100 000 people. However, more than 3 million tonnes of plastic waste are disposed of in Canada every year. Despite having extensive recycling programs, only 9% of plastic materials are recycled in Canada, with a large proportion ending up in landfills or the natural environment (
Environment and Climate Change Canada 2023). Moreover, ∼80%–90% of the solid waste found in marine environments is plastics of different sizes (
Environment and Climate Change Canada 2023). Plastic disposal has a significant impact on human and ecological health, either directly or indirectly (
Ali et al. 2021b). For example, toxic microparticles and nanoparticles are generated when polystyrene is degraded in the natural environment by either exposure to the UV radiation of the sun or physically by wind and water (
Andrady 2011;
Gewert et al. 2015). These micro/nano particles may induce inflammation in tissues and organs as well as hemolysis (destruction of red blood cells) (
Hwang et al. 2020).
The most widely used plastics include polyethylene (PE), polypropylene (PP), polyvinyl chloride (PVC), polyethylene terephthalate (PET), and polystyrene (PS) (
Andrady 2011). PS is a synthetic aromatic polymer made from styrene monomers with the molecular formula of (C
8H
8)
n. The four main types of PS include general purpose PS (GPPS), high impact PS (HIPS), polystyrene foam (PF), and expanded polystyrene (EPS) foam, often referred to as Styrofoam (
American Chemistry Council 2015;
Ho et al. 2018).
PS disposal is challenging because it does not break down easily and tends to persist in the environment. In general, one of the main degradation mechanisms of synthetic polymers is through enzymatic hydrolysis (
Ali et al. 2021a,
2021b). Some synthetic plastics, such as polyurethane (PU), have a C–O backbone that makes them more susceptible to enzymatic degradation. However, PS consists of a C–C backbone, which is generally difficult to break down via hydrolysis (
Krueger et al. 2015;
Zhang et al. 2022). In the natural environment, plastic degradation starts with photodegradation mainly via exposure to the UV radiation of the sun.
Since landfill disposal of PS is not a long-term sustainable solution, alternative options are needed to reduce PS quantities that end up in landfills and the natural environment. Recent studies have reported various invertebrate organisms that could enzymatically degrade PS. These include yellow mealworms,
Tenebrio molitor L. (
Yang et al. 2015a,
2015b;
Brandon et al. 2018;
Yang et al. 2018a,
2018b), and superworms, such as
Zophobas atratus (
Peng et al. 2019,
2020).
It has been reported that the microorganisms present in the gut of some insect larvae contribute to the degradation of PS (
Brandon et al. 2018). For example, removal of gut bacteria via antibiotic treatment reduced biodegradation efficiency in superworms (
Zophobas atratus) (
Yang et al. 2020).
Zophobas atratus is a heterotypic synonym of
Zophobas morio. A variety of bacteria contribute to PS biodegradation in the guts of superworms. For example,
Sphingobacterium spp. and
Dysgonomonas spp
. were found in the gut of PS-fed superworms (
Luo et al. 2021).
Pseudomonas spp
. from superworms have also been shown to degrade PS (
Kim et al. 2020). However, little information exists on the conditions that can enhance bacterial degradation, as well as the nature of degradation products, and whether these products could be used as fertilizers.
Several studies have investigated various methods to enhance degradation of PS. For example, a study has compared enzymes from various bacterial species for PS degradation (
Mohanan et al. 2020). Others have explored the use of physical methods, such as UV radiation (
Chamas et al. 2020), or chemical additives, such as starch, to improve PS degradation (
Mohanan et al. 2020). However, studies on the potential use of superworms for PS degradation, as well as the characteristics of their degradation products, are scarce.
The objectives of this laboratory-based study were to evaluate nutrient (nitrogen and phosphorus) content and degradation product composition of frass and to identify the gut bacteria in superworms (Zophobas morio) following the ingestion of EPS.
Materials and methods
Experimental design and set-up
The study was conducted in a laboratory using a two-factor experimental design with four replicates per treatment to give a total of 24 experimental units. The two factors studied were as follows: Factor 1 was the type of diet fed to the superworms, which was either S-shaped EPS chips or block-type EPS or wheat bran as the control. The S-shaped EPS is typically anti-static and of lower density (3.8– 4.0 kg/m3), whereas the block-type EPS is typically higher density (14–16 kg/m3). Factor 2 was the type of pre-treatment applied to the polystyrene samples, which was either a 60-second direct exposure to UV radiation using a transilluminator (Fisher Scientific Model FTBI-614 312 nm with total UV intensity of 8000 µw/cm2) or no UV radiation. The UV treatment time of 60 s was selected because it was short enough to minimize exposure to surrounding environment, cost-effective if this could be adopted by waste management facilities, and potentially long enough to loosen bonds with the polystyrene structure and make it easier to break down. The mass of EPS used in each container was 1.44 g.
The superworms were initially fed a wheat bran diet before the experiment started and were then starved for 48 h before being fed only S-shaped EPS chips, block-type EPS, or wheat bran (control). A total of 12 superworms were placed in each 500 mL plastic beaker and covered with perforated parafilm to prevent superworms from escaping. The study was conducted at a room temperature, approximately 19–20 °C. Diet treatments were sprayed with deionized water once a day. No nutrients were added to either treatment.
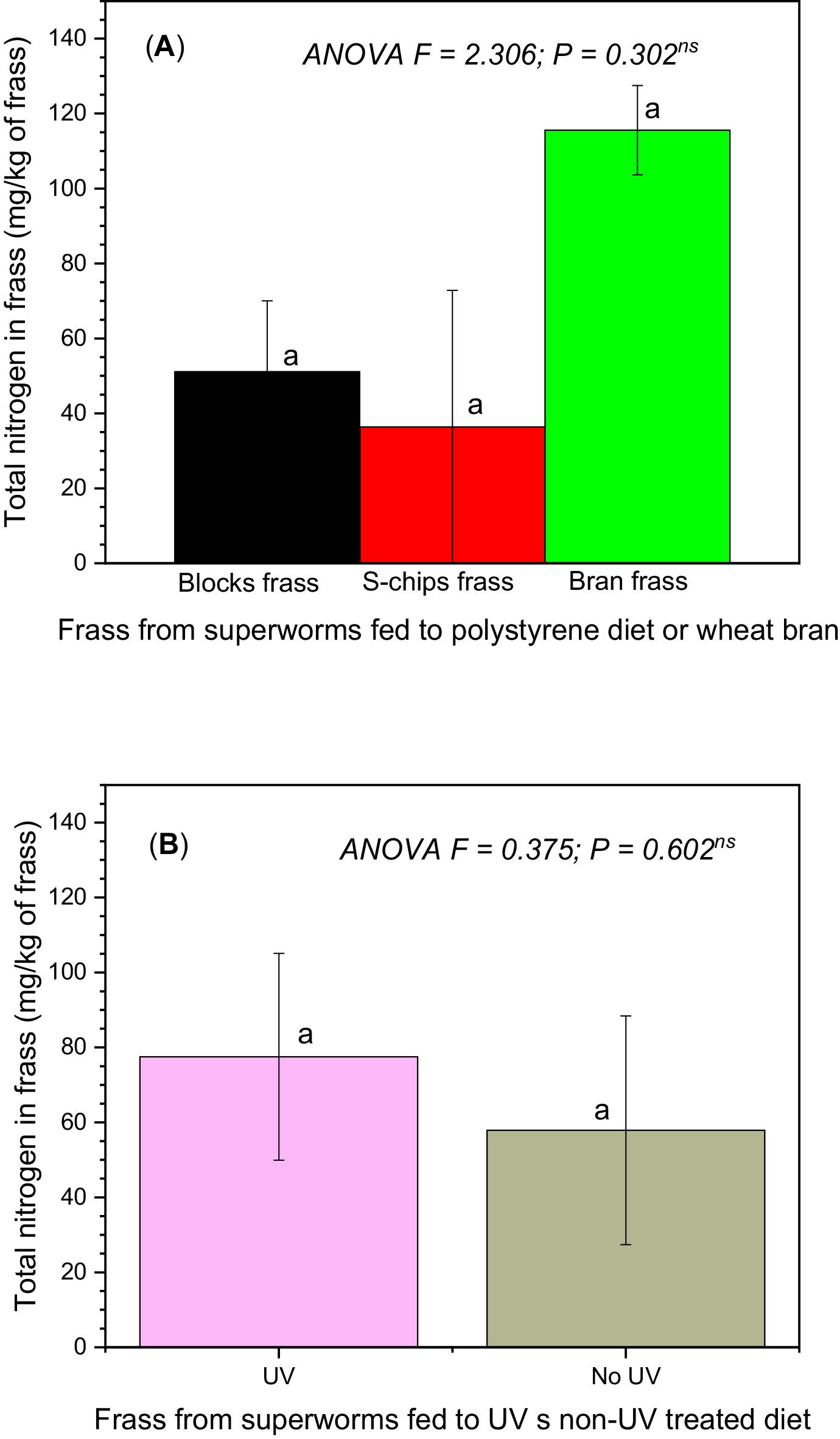
Following a 28-day incubation period, EPS loss of mass was measured, and the frass of superworms was collected and analyzed. Total nitrogen and phosphorus contents of frass were determined using UV–visible spectrophotometry.
Loss of mass and survival rates
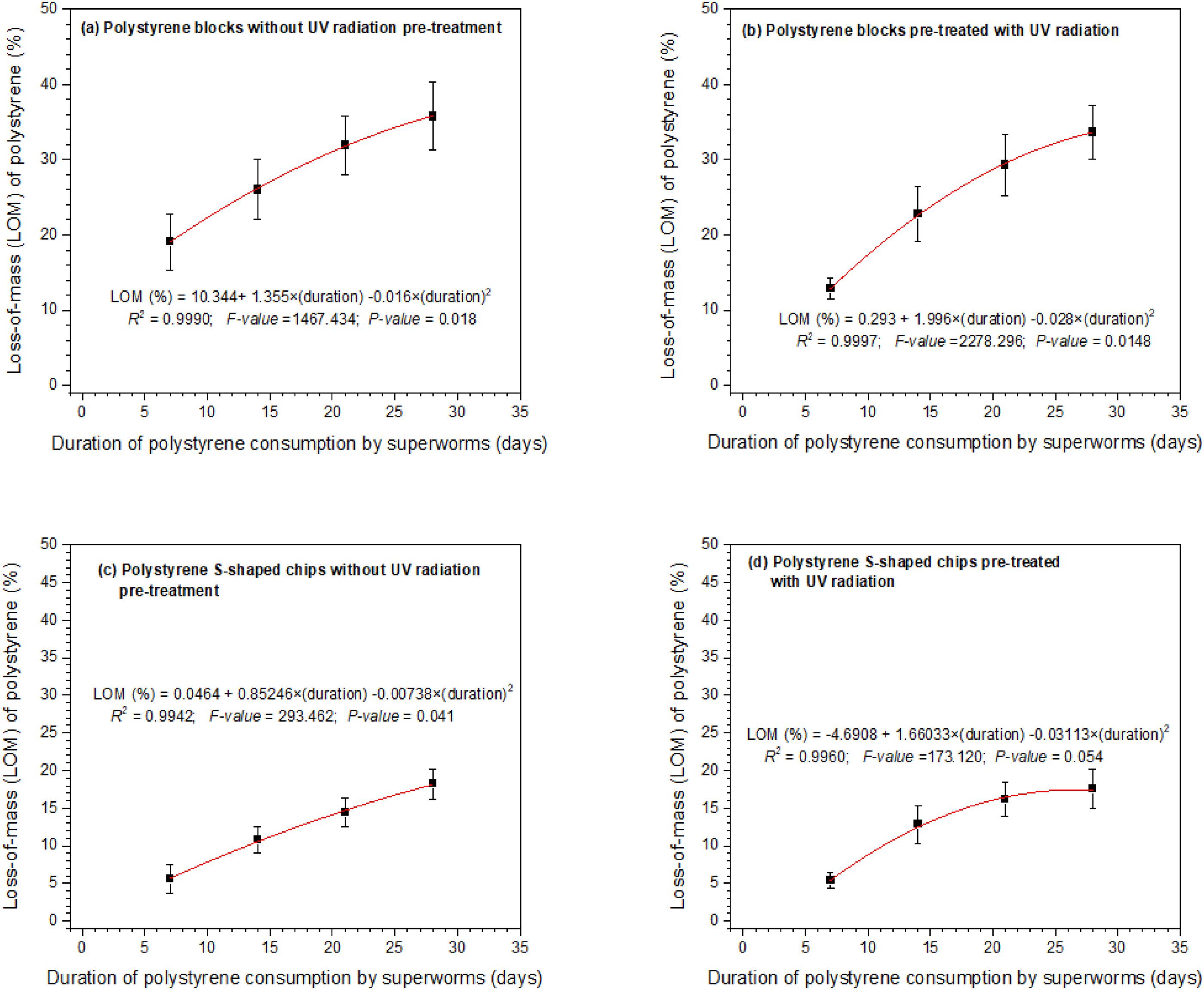
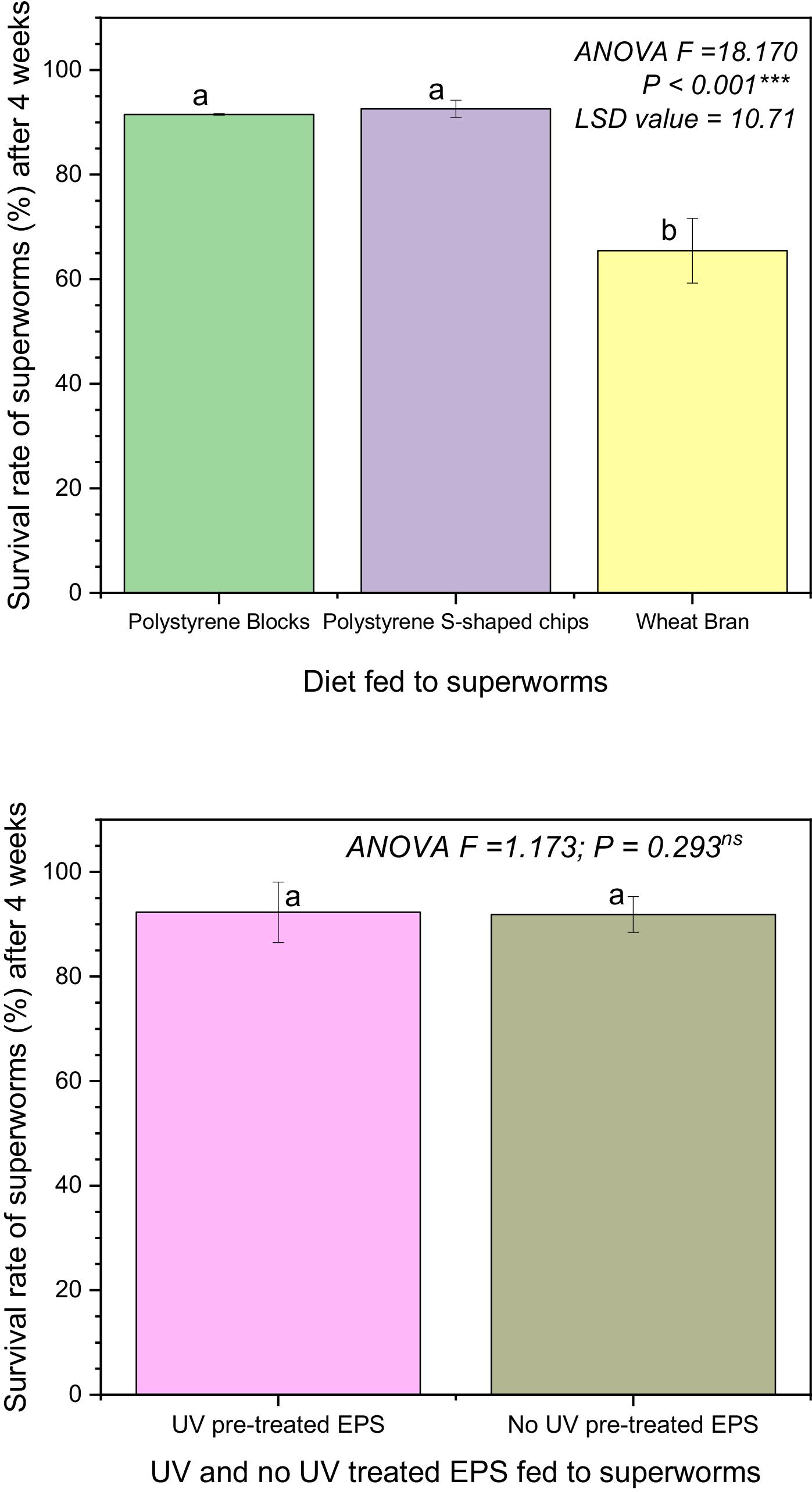
The loss of mass measurements (in grams) were conducted on a weekly basis for 4 weeks. It involved carefully removing the intact EPS blocks or S-shaped chips in each container and weighing them. The small EPS particles resulting from superworms eating the chips or blocks were not weighed as these were scrapped off using a small brush. The percent loss of mass at the end of each week was calculated by dividing the loss-of-mass of that week by the original mass of EPS in each container (i.e., 1.44 g). The loss of mass rate (in mg per day per superworm) was calculated at the end of the 28-day period for each container by dividing the total loss of mass in that container by 28 and then by the number of superworms that survived the experiment. Survival rate was determined as the total number of superworms that survived each week until 28 days of feeding on one of three diets divided by the number of superworms at the beginning of the feeding period and was expressed as a percentage.
Statistical analysis
Statistical analysis was conducted using the Statistical Package for Social Sciences (SPSS) Version 26. The Analysis of variance (ANOVA) was conducted using the general linear model procedure to assess the data of each week. The sources of variation included in ANOVA were the type of diet, the type of pre-treatment applied to the polystyrene samples, and the potential interaction between them. Statistical significance was determined when P value (ANOVA) < 0.05.
Preparation of frass samples for chemical analysis
Different sieve sizes (0.5 and 1.0 mm) were used to separate large EPS particles from the frass. The final separation was conducted by blowing air gently in a receiving pan. The frass from all four replicates per treatment were pooled prior to chemical (i.e., nitrogen, phosphorus, and degradation products) analysis.
Nitrogen analysis
Nitrogen extraction of frass was performed using modified Kjeldahl method (
APHA 1999) in three steps: digestion, distillation, and titration. A precisely weighed frass sample (∼0.1 g) was placed into a Kjeldahl flask. A total of 6 mL of H
2SO
4 and 2.89 g of K
2SO
4 were added to the flask plus a small piece of copper as the catalyst to increase the reaction rate of converting organic nitrogen into ammonium and therefore reduce the reaction time. The flask was then heated up to 370 °C for 75 min.
Distillation was conducted after the solution cooled down to room temperature. A total of 100 mL of deionized water was added to the flask. The digest was made basic by adding approximately 25 mL of 45% w/v NaOH to the flask to convert ammonium (NH4+) into ammonia gas. Ammonia (NH3) was then steam distilled into a solution made of 28 mL distilled water mixed with 6 mL of 12 mol/L HCl. The solution was then titrated with standardized 1 mol/L NaOH solution to the colorless end point using phenolphthalein as indicator.
Phosphorus analysis
Phosphorus determination was conducted using the modified rapid digestion method by
Pequerul et al. (1993). A total of 0.25–0.30 g of the air-dried frass sample was digested in 5 mL of 15% H
2O
2 and 5 mL of concentrated (15.8 mol/L) HNO
3. The digest was cooled down, syringe filtered (2.5 µm), washed with 5 mL of 6 mol/L HCl, and centrifuged (269 ✕
g, 10 min). The supernatant was then diluted to 25 mL using chromatography-grade water and acidified with 5 mL of 1:1 HCl, to ensure that final pH was much lower than 7. Phosphorus content was determined in the supernatant (10 mL) using the ammonium molybdate/stannous chloride method (
APHA 1999) with absorbance measured at 690 nm.
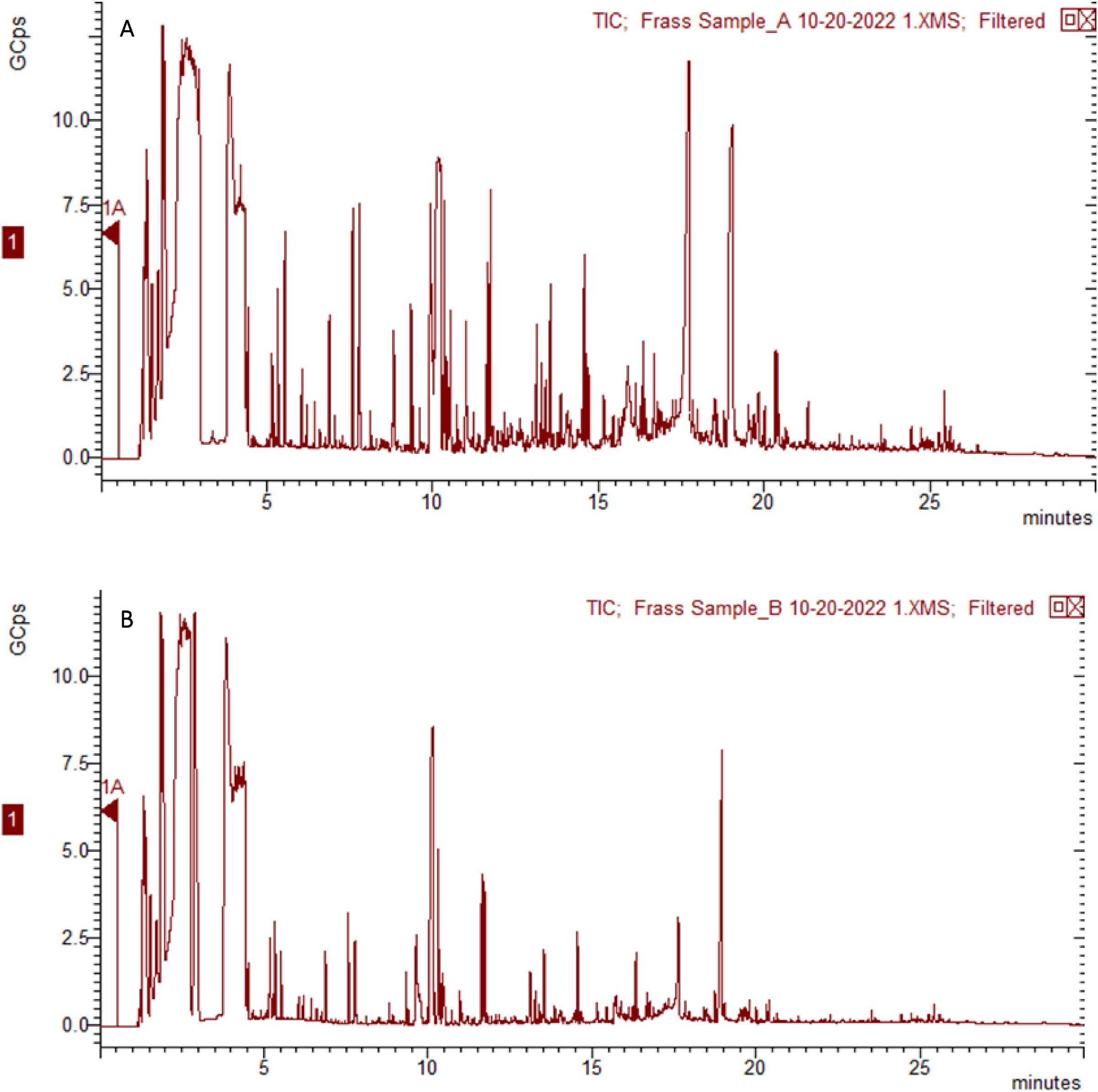
Analysis of polystyrene degradation products in frass
Five hundred microliters of 50% methanol in chromatography-grade water were added to 100 mg of frass and mixed vigorously (3 ✕ 20 s) then centrifuged (15 000 ✕
g, 15 min, 4 °C). The supernatant (200 µL) was filtered (0.46 µm) and dried under nitrogen (30 °C). Extracted compounds were derivatized with bis(trimethylsilyl)trifluoroacetamide (85 µL, 80 °C, 1 h) and analyzed by a Bruker Scion 436 TQ Gas Chromatography-Mass Spectrometer (GC-MS/MS) instrument on a HP-5MS 30 m ✕ 0.250 mm ✕ 0.25 µm column (Agilent Technologies) using an adaptation of a published method (
Tsochatzis et al. 2021). Briefly, derivatized compounds (1.0 µL) were injected into the GC column at a 10:1 split ratio with an injector temperature of 300 °C. The oven temperature was held at 60 °C for 2 min, then raised to 290 °C at a rate of 10 °C/min, and held at 290 °C for 5 min. Full-scan MS analysis was performed for
m/z 50–600 with transfer line temperature and source temperature both set to 290 °C. The total analysis time was 30 min. Major GC-MS peaks (>2 GCps) were extracted and their MS spectra were searched against the Human Metabolome Database (HMDB) version 5.0 using its GC-MS search tool with MS tolerance set to ±0.5 Da (
Wishart et al. 2022). Only compounds with a proportion of the library spectrum’s peaks with matches > 0.80 were considered. A library match score > 0.80 means at least 80% of the MS peaks obtained from the sample were identical to those in the library spectrum i.e., compound was a “match”. Unusually derivatized (such as trimethylsilylated compounds) and physiologically irrelevant compounds (i.e., compounds not supposed to be found in the sample, such as drugs or household products) were excluded from the search.
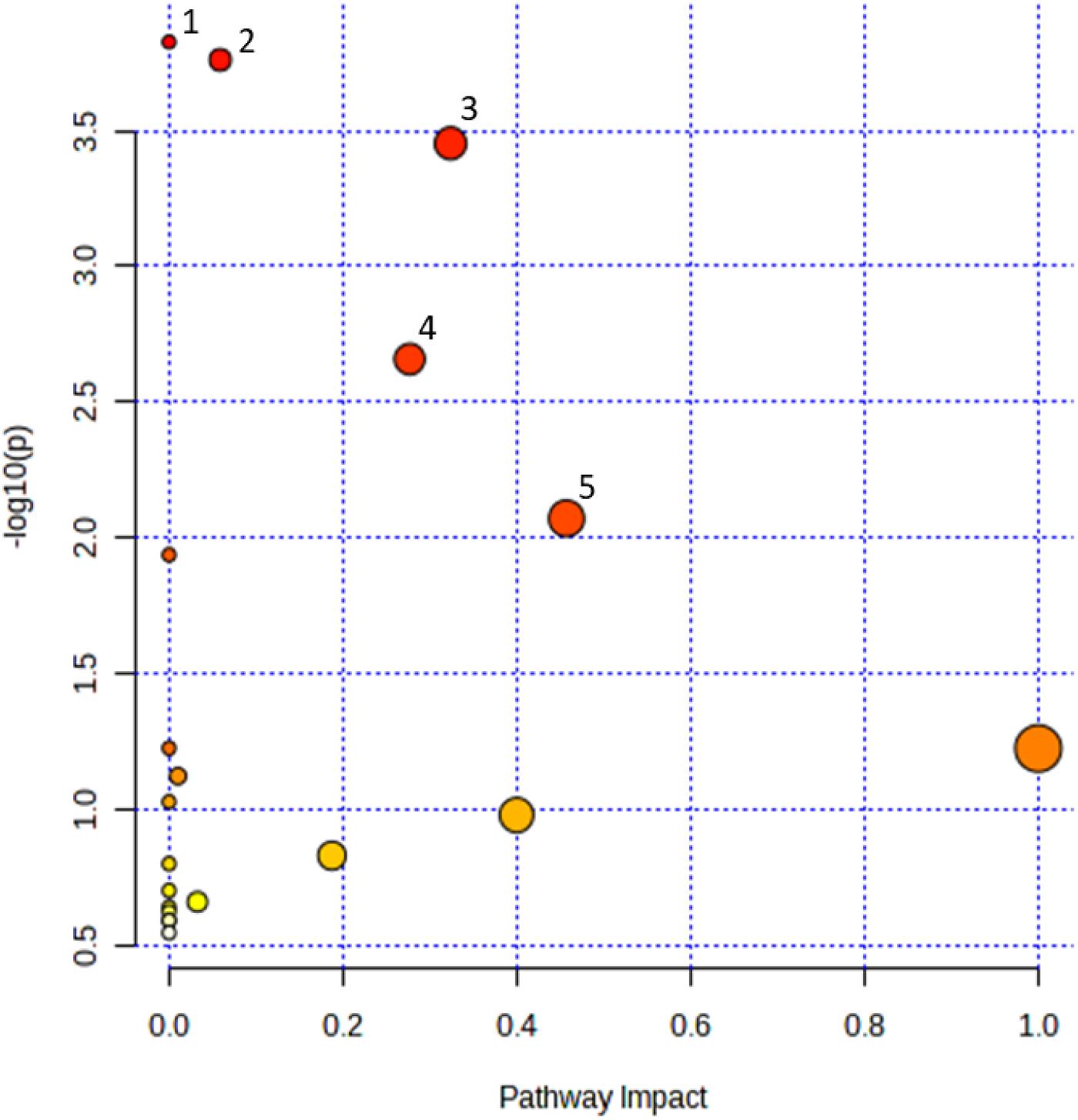
Metabolic pathway analysis
Pathway analysis was performed on the identified HMDB hits using the MetaboAnalyst 5.0 web tool for metabolomics data analysis (
Pang et al. 2022) with hypergeometric testing and relative betweenness centrality set for enrichment and topology analyses, respectively, in
Drosophila melanogaster as the closest species in the web tool to
Zophobas morio. Significantly altered pathways were visualized on a scatter plot with pathway impact and ‒log
10p on the
x- and
y-axes, respectively. Pathway impact was calculated by the web tool as the sum of importance measures of each of the matched metabolites divided by the sum of the importance measures of all metabolites in each pathway.
Preparation of polystyrene film
To form polystyrene films, 1.5 g of EPS pieces were dissolved in xylene and the solution was left to dry for 24 h. The film was removed, washed with 10 mL of 100% methanol and then rinsed with deionized water (DW), and then left to dry for three more days.
Liquid carbon-free media
Liquid carbon-free medium (LCFBM) was prepared by dissolving 0.7 g of KH2PO4, 0.7 g of K2HPO4, 0.7 g of MgSO4·7H2O, 1.0 g of NH4NO3, 0.005 g of NaCl, 0.002 g of FeSO4·7H2O, 0.002 g of ZnSO4·7H2O, and 0.001 g of MnSO4·H2O in 1 L of DW. Luria-Bertani (LB) broth medium was prepared by dissolving 25 g of LB Miller broth into 1 L of DW. To make the solid version, 15 g of agarose was added to the medium. All the media were autoclaved at 121 °C for 15 min.
Extraction of gut bacteria
Bacteria were extracted from larvae that had been fed EPS for 28 days as their sole diet. The larvae were submerged in 70% ethanol for 3 min. After submersion the larvae were dissected, and three larval guts were placed in a 15 mL tube containing 3 mL of liquid carbon free media. The tubes were shaken for 15 min and the gut tissue was removed. The tubes were centrifuged (448 ✕ g, 5 min) at room temperature to obtain a pellet followed by removal of supernatant and pellet resuspension in 5 mL of LCFBM. The washing step was repeated two more times with the final pellet being resuspended in 1 mL of LCFBM.
Isolation and identification of bacteria
The collected gut cell suspension was used to inoculate liquid cultures containing 0.50 g of sterile EPS pieces or EPS film in a 250 mL Erlenmeyer flask (n = 4) with 100 mL of LCFBM. The cultures were incubated at 24 °C and 120 RPM for 10–11 days of growth; dilutions of 1/10, 1/100, and 1/1000 of the liquid cultures were plated onto LB or bovine heart infusion for colony isolation.
Colonies from the plates were selected based on colony morphology (shape, colour, and size). Each colony was added to a 5 mL culture tube with 2 mL of LB and incubated at 24 °C and 120 RPM for 24 h.
To establish that the isolated bacteria were able to grow on this media as an individual species, an inoculum from each colony was added to LCFBM containing EPS. Samples were taken at regular intervals and examined by spectrophotometer to assay for increased numbers of cells.
DNA extraction and amplification
Bacterial DNA was isolated using a Blood and Tissue kit from Qiagen following the instructions for gram-positive cells included in the kit. The concentration of DNA was determined by a nanodrop spectrophotometer. For those bacteria that displayed degradation capabilities, their 16S rRNA gene was amplified by PCR targeting the hyper-variable V4 region using the following primers:
•
forward primer 515 F (5′-GTGCCAGCMGCCGCGGTAA-3′)
•
reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′).
The final volume of this PCR was 50 µL, with 12.5 µL trehalose (20%), 5 µL buffer, 3 µL MgCl2 (50 mM), 1 µL dNTPs (10 mM), 2.5 µL forward primer (10 µmol/L), 2.5 µL reverse primer (10 µmol/L), 1 µL Taq polymerase (5 U/µL), and 100 ng of template DNA. PCR was performed under the following conditions at 94 °C for 2 min, 94 °C for 30 s, 56 °C for 40 s, 72 °C for 1 min, 72 °C for 10 min. The PCR products were sent to Genome Quebec for 16S rRNA gene sequencing. The returned sequences were Basic Local Alignment Search Tool (BLAST) searched in the NCBI database and compared to the observed bacteria characteristics to confirm identification.
Discussion and conclusions
The loss-of-mass rates per day per superworm observed in our study was within the same range reported for mealworm larvae (
Tenebrio molitor L.) which ranged between 0.04 and 0.4 mg per day per mealworm observed in related studies (
Brandon et al. 2018;
Yang et al. 2018a;
Yang et al. 2021;
Palmer et al. 2022). More specifically the loss-of-mass rate for the EPS fed to mealworms over a 32-day period averaged 0.10 mg per day per mealworm (
Yang et al. 2021), which was equal to what we also observed in our study for superworms fed on expanded EPS over a 28-day period. In a separate study
Yang et al. (2020) observed that superworms (
Zophobas atritus) consumed EPS at a rate of 0.58 mg per day per superworm, which was four times higher than consumption rate by mealworms (
T. molitor). That study also reported that the superworms were able to live normally on an EPS diet for 28 days.
The decrease in the mass of EPS S-shaped chips and white blocks over the 28-day period was significant. The decrease over time was relatively consistent, regardless of whether the polystyrene had been pre-treated with 60 s of exposure to UV radiation.
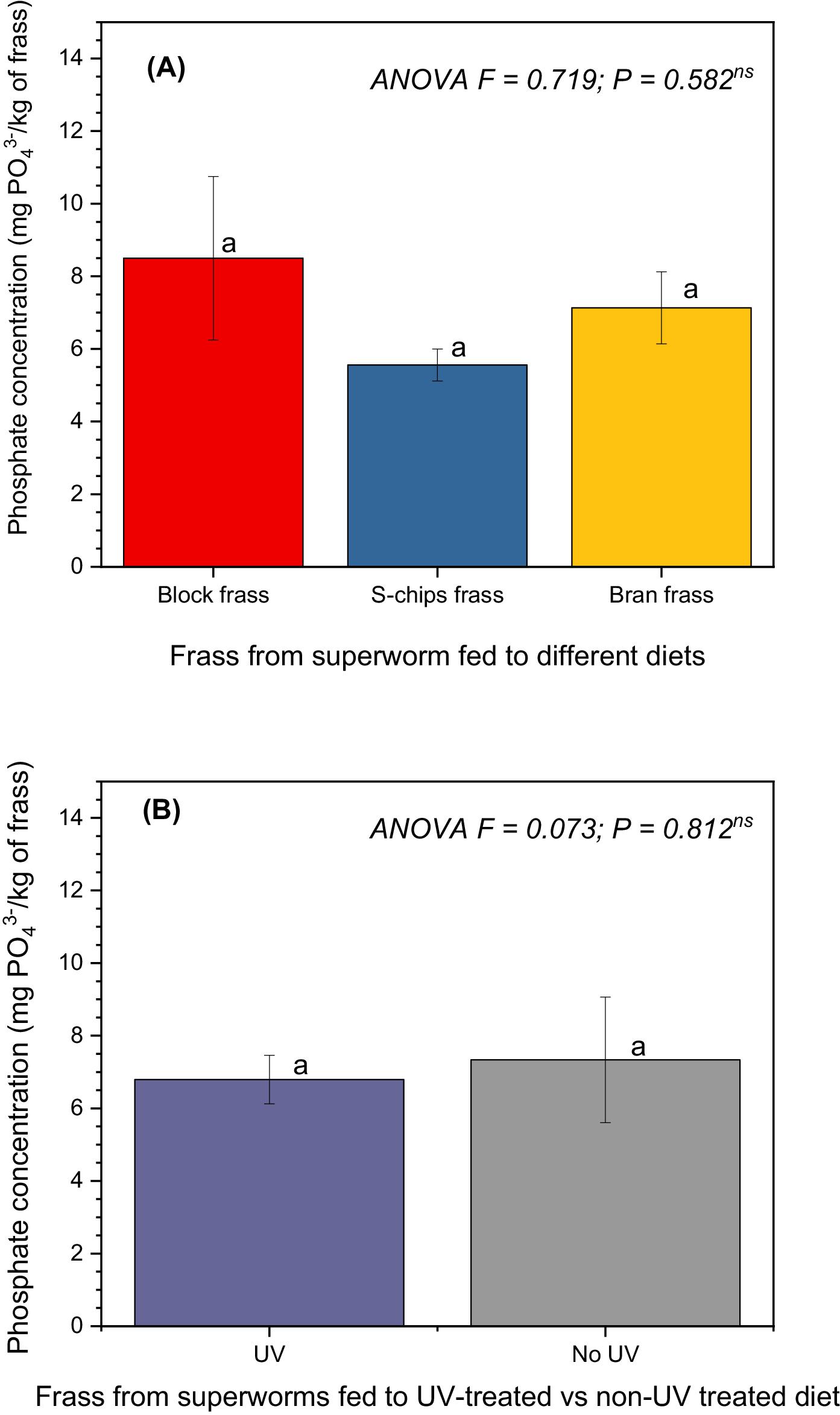
The phosphorus content of 1.8–2.8 g/kg frass observed in our study is much lower than that reported for insect frass contents of up to 20 g/kg frass (
Houben et al. 2020). However, in our study regardless of whether the superworm larvae fed on EPS diets (S-shaped chips and white blocks) or wheat bran, the total P content was not different, indicating that this could be perhaps the naturally low P content found in superworm frass. Although the specific details of the composition of EPS used in this study are not available, it is common for PS manufactured to have additives that contain nitrogen and phosphorus. Four additives commonly used in PS production and have significant nitrogen content include 2-(2′-hydroxy-5′-methylphenyl)benzotriazole) (Tinuvin P), bis (2, 2, 6, 6-tetramethyl-4-piperidyl) sebacate (Tinuvin 770), which are both used in PS production as UV absorbers to stabilize PS when exposed to UV radiation, N, N-ethylene-bis(tetrabromophthalimide) (Saytex BT-93) which is used as flame retardant, and a N, N’-ethylene-bis-stearamide-based synthetic wax (Acrawax) used as a lubricant (
Ho et al. 2018). These four additives are used at rates of 0.2%–0.3% for Tinuvin P, 0.2%–0.5% for Tinuvin 770, up to 13% for Saytex BT-93, and 200 ppm for Acrawax, and therefore likely provided a source of nitrogen for the superworms feeding on the EPS diet. Thirteen of the 21 degradation products that we identified in frass were nitrogen-containing molecules, including an amine diol, an aromatic amine, 2 aliphatic diamines, and 7 amino acids.
Another additive used in PS production is tris(nonylphenyl) phosphite (Wytox) that contains phosphorus, and its antioxidant properties reduce the degradation of PS (
Ho et al. 2018). These compounds can be used in PS production at amounts of 0.2%–0.3% for Tinuvin P and 0.2% for Wytox (
Ho et al. 2018). Therefore, the phosphorus observed in our frass samples may have originated from PS additives used during its manufacture. Part of this phosphorus was found to be biotransformed into phosphoric acid and pyrophosphoric acid.
Our results showed that the pre-treatment of EPS with UV radiation exposure for 60 s did not result in significant differences in loss of mass, nitrogen content in frass, and phosphorus content in frass. However, degradation products identified from GC-MS analyses of frass showed more abundance of amino acids and short-chain carboxylic acids in frass from superworms fed on EPS diet. Pre-treatment with 60 s of UV radiation exposure resulted in high quantities of degradation products compared to non-UV treated frass samples. A study by
de Castro Monsores et al. (2021) showed that UV pre-treatment of PS resulted in modification of the macromolecules which affected its mechanical properties, such as scission of molecular chains. Other studies using various organisms have confirmed that UV radiation pre-treatment can enhance degradation of plastics. For example,
Esmaeili et al. (2013) reported that exposure of PE to
Aspergillus niger fungal species for 126 days resulted in degradation percentages of 30% for UV treated versus 16% for non-UV treated PE. Similarly enhanced degradation of UV pre-treated polypropylene (PP) by
Phanerochaete chrysosporium NCIM 1170 (F1) and
Engyodontium album MTP091 (F2) fungi was also reported (
Jeyakumar et al. 2013). Similar to what has been reported for plastic degradation in natural environments, the UV pre-treatment of PS initiates photo-oxidation that causes cleavage of polymer bonds resulting in shorter chain oxygen containing products that are more prone to biodegradation by microorganisms (
Yousif and Haddad 2013).
Some studies have reported that frass from superworms fed to polystyrene diets had high content of amides (
Gan et al. 2021). However, we did not identify any amides among the most abundant compounds in our frass samples. A study by
Jiang et al. (2021) identified several alcohols and carboxylic acids in the gut and frass of superworms that had fed on PS diet, including phthalic acid, which was identified in our analysis as well. However, compared to the aforementioned study, we identified carboxylic acids with a shorter carbon chain, such as succinic acid, glycolic acid, and lactic acid.
Chemical characterization studies have reported total nitrogen contents of up to 50 g/kg frass (i.e., equal to 5% or 5000 mg/kg frass) from mealworms that had exclusively been fed to wheat bran diet (
Houben et al. 2020).
Previous studies (
Peng et al. 2020,
2022a,
2022b) have used gel permeation chromatography, coupled with Fourier-transform infrared spectroscopy to monitor the process of PS degradation in mealworm and superworm frass. We chose to use GC-MS as it allowed us to also characterize the degradation products.
Our results show that PS exposure has triggered several metabolic pathways in superworms. A study by
Peng et al. (2022b) equally showed many metabolic pathways found in mealworm (
Tenebrio molitor) gut. Among these pathways, the pathways that overlapped between the two studies are arginine and proline metabolism, D-glutamine and D-glutamate metabolism, and histidine metabolism.
No previous studies have reported the identification of amino acids in the frass of superworms.
A study found that some species, such as larval zebrafish, exhibited increased levels of some amino acids such as glutamine, beta-alanine, or some acids such as methylmalonic acid and succinic acid, while also decreasing in the levels of putrescine in the presence of PS (
Wan et al. 2018).
The gut microbial community of superworms is thought to contribute to the degradation of EPS. In our study superworm gut bacteria were isolated to identify species that may be responsible for EPS degradation. The isolates were grown on media containing EPS as the sole carbon source and DNA from the isolates was extracted, then the V4 region of the 16S rRNA gene was amplified by PCR and sequenced. One of sequences analyzed by BLAST was identified to belong to
Stenotrophomonas sp. The genus
Stenotrophomonas is composed of at least sixteen species that have been identified from various sources (
Deng et al. 2022). The main environmental reservoirs of the
Stenotrophomonas spp. are plants and soil. These gram-negative bacteria are mainly non-pathogenic except for the opportunistic pathogen
Stenotrophomonas maltophilia. Most species of
Stenotrophomonas promote plant growth via several mechanisms including nitrogen fixation (
Park et al. 2005), solubilization of phosphate, as well as protecting the plant from pathogens and promoting growth under stressing conditions (
Ryan et al. 2009). Members of this genera have been found to persist in the gut of the bark beetle
Dendroctonus rhizophagus throughout its life cycle (
Morales-Jiménez et al. 2012).
To the best of our knowledge
Stenotrophomonas sp. has not previously been identified as a component of the gut microbial community in superworm larvae. This genus of bacteria has been identified from the gut of meal worms that have been fed an exclusively EPS diet and a polyethylene diet that included bran (
Lou et al. 2021).
Stenotrophomonas sp
. isolated from solid waste dump sites has been shown to degrade low-density polyethylene (
Nadeem et al. 2021).
Stenotrophomonas produces polyamines like putrescine and cadaverine that correlate with root development and could protect the plants from environmental stresses (
Ulrich et al. 2021). Decarboxylation of ornithine or arginine is the first step in the synthesis of polyamines (
Ulrich et al. 2021). It is interesting to note that polyamines and arginine were found among the degradation products tentatively identified in EPS-fed superworm frass
Table 1.
We also identified
Pseudomonas sp. as constituents of the superworm gut. A study by
Kim et al. (2020) reported that of all the gut bacteria identified in superworms using DNA sequencing analysis, 35% were
Pseudomonas aeruginosa. Our study identified
Pseudomonas aeruginosa from superworm guts, the same bacteria species that has also been identified by other researchers in superworm guts (
Kim et al. 2020;
Lee et al. 2020;
Li et al. 2020).
Pseudomonas aeruginosa isolated from guts of superworms has been found to degrade PS at a rate of 0.1% per day, and the serine hydrolase enzyme secreted by this bacteria species was involved in PS degradation (
Lee et al. 2020).
In conclusion, our study results showed significant loss-of-mass of EPS blocks and S-shaped chips after 28 days of consumption by superworms. Survival rates of superworms that relied on EPS diet alone were greater than 90% for both EPS blocks and S-shaped chips. Pre-treatment using UV exposure for 60 s did not affect loss of mass rates for EPS, or nitrogen and phosphorus content in frass. Frass from the superworms contained nitrogen and phosphorus, and therefore could potentially serve as a fertilizer. GC-MS analyses of frass showed that UV pretreatment resulted in higher peak areas and more complex degradation products that included amino acids and short-chain carboxylic acids. Gut bacteria isolated from the superworms using PCR amplification and 16S rRNA gene sequencing followed by BLAST search showed sequence identity with Pseudomonas aeruginosa and Stenotrophomonas sp. Our study findings suggest that superworms have the potential to degrade polystyrene, however further research is needed to determine whether this could be a sustainable approach to managing polystyrene.