A combination of optimized threshold and deep learning-based approach to improve malaria detection and segmentation on PlasmoID dataset
Abstract
Malaria is a life-threatening parasitic disease transmitted to humans by infected female Anopheles mosquitoes. Early and accurate diagnosis is crucial to reduce the high mortality rate of the disease, especially in eastern Indonesia, where limited health facilities and resources contribute to the effortless spread of the disease. In rural areas, the lack of trained parasitologists presents a significant challenge. To address this issue, a computer-aided detection (CAD) system for malaria is needed to support parasitologists in evaluating hundreds of blood smear slides every month. This study proposes a hybrid automated malaria parasite detection and segmentation method using image processing and deep learning techniques. First, an optimized double-Otsu method is proposed to generate malaria parasite patch candidates. Then, deep learning approaches are applied to recognize and segment the parasites. The proposed method is evaluated on the PlasmoID dataset, which consists of 468 malaria-infected microscopic images containing 691 malaria parasites from Indonesia. The results demonstrate that our proposed approach achieved an F1-score of 0.91 in parasite detection. Additionally, it achieved better performance in terms of sensitivity, specificity, and F1-score for parasite segmentation compared to original semantic segmentation methods. These findings highlight the potential of this study to be implemented in CAD malaria detection, which could significantly improve malaria diagnosis in resource-limited areas.
1. Introduction
Patients suffer from malaria when their blood contains malaria parasites. The parasites are transmitted into the body through a bite of infected female Anopheles mosquitos, and they multiply in the red blood cells (RBCs) and spread to other cells. According to the World Health Organization (WHO), malaria is a public health issue that spreads to most countries, mainly tropical countries. Up to 90 countries were endemic malaria areas and more than four hundred thousand dead people in 2017. The most vulnerably infected, 61%, were children under 5 years (Anon 2018).
One of the most extensively used malaria diagnosis techniques is the malaria rapid diagnostic test. It provides less than a 10% false positive (FP) rate. In addition, the test is easy to use and offers quantitative diagnoses within <30 min. However, this technique cannot determine the number and type of parasites (Moody 2002).
Another malaria diagnosis technique, the cornerstone of parasite-based malaria diagnosis, is a manual light microscopic examination. This technique encourages further analysis by observing a micron part on the slide using a zooming microscope. However, the diagnosis result of this technique depends on the examiner's expertise and thoroughness. In addition, this technique is laborious because the examiner usually takes 30 min to find and count the parasite in a single thin blood smear (Maqsood et al. 2021).
Hundreds of blood smear films are examined every week in the endemic area. Therefore, it carries massive resources and economic demand (Maqsood et al. 2021). Moreover, some previous studies found that manual microscopy assessments produced subjective results due to different experience of the examiner (Thimasarn et al. 2002; Mitiku et al. 2003; Bates et al. 2004; Tek et al. 2009). This situation is even worse because of lack of experienced examiners in the rural areas. Furthermore, the lack of health facilities and resources in rural areas might negatively affect blood smear film quality, as there are many presented artifacts and noises. Automated malaria detection methods use minimum human intervention. Therefore, the automated detection systems are more objective, reliable, and efficient than manual examinations.
Today, many automated malaria parasite detection methods with excellent performances have been proposed. Generally, there are three groups of methods for automating malaria parasite detection, namely, traditional methods, modern (artificially intelligent) methods, and hybrid methods. The traditional methods involve human knowledge in the malaria parasite detection process. The traditional methods mostly use complex image processing techniques to produce the object candidates and extracting object features using hand-engineered features, namely, color, intensity, shape, and texture (Frean 2010; Moon et al. 2013; Poostchi et al. 2018; Nugroho 2019a, 2019b). There are three main steps in the traditional method. The first step is to segment RBCs from microscopic thin blood smear images using various image segmentation techniques. The second step is to compute a set of features applying hand-engineered techniques, and then feed these features into a classifier. The final step is to classify the infected RBCs.
Several public datasets have been introduced to support the development of automated malaria detection and segmentation systems. Quinn et al. (2014) presented a malaria public dataset. The dataset included 1182 microscopic images of thick blood smears captured by a smartphone camera. It consisted of 948 malaria-infected images with 7628 parasites. However, the parasites only consist of Plasmodium falciparum. An early-generation malaria dataset was introduced by Loddo et al. (2018). The dataset consisted of 229 microscopic images captured from thin blood smear films using Leica optical laboratory microscope. It contained 483 malaria-infected blood cells of 48 000 blood cells. The dataset included all malaria parasite species and their life stages. However, the parasite class distribution is significantly unbalanced. One of the classes consists of 695 parasites, and the other four classes consist of less than five parasites. The other large malaria dataset is introduced by Sultani et al. (2022). They introduced a malaria dataset captured with multimagnification scales in 1257 thin blood smears with 3624 parasites. However, this dataset only contains Plasmodium vivax. Recently, Nugroho et al. (2022) presented a new dataset for malaria parasite detection and segmentation in thin blood smears. This dataset includes 559 microscopic images with 691 malaria parasites. It was captured from hundreds of thin blood smears collected from rural areas in Indonesia. The advantage of this dataset is that it includes all malaria parasite species and their life stages with better class distribution than Loddo et al. (2018).
Maysanjaya et al. (2016) applied Otsu method to find the global threshold value for segmenting P. Vivax, one kind of malaria parasite. They used a combination of the red channel of RGB and the saturation channel of HSV color space as pre-processing to suppress the noise. The method performance achieved an accuracy of 0.93. Dave and Upla (2017) also applied the original Otsu technique to find the global threshold value for RBCs. In addition, they used some rule-based methods to segment the parasites. Unfortunately, this study does not state the malaria parasite types in their dataset. However, both these studies use a dataset containing only 30 microscopy images with well-separated RBCs. To find a global threshold value, Memeu et al. (2013) proposed a double Otsu method to segment the dark objects, such as parasites, white blood cells (WBCs), and artifacts. The segmented areas were then classified by an artificial neural network to find the parasites. As a result, they obtained an accuracy of 0.95. Moreover, the proposed method achieved a fast implementation in computational time.
Currently, deep learning (DL) approaches have successfully eliminated the issues of traditional methods, such as hand-crafted feature extraction, unreliability, and complex rule base. Instead, DL models have hidden layers whose task is to extract the features automatically (Nautre et al. 2020). However, DL approaches need an extensive dataset for training their model to obtain a good performance. In addition, collecting datasets in the medical field is more difficult and limited compared to non-medical fields. Therefore, many studies have introduced some image augmentation algorithms to overcome the problem of small data or even imbalanced data. The algorithms are also based on image processing and DL techniques. Traditional image augmentation methods based on image processing include geometric transformations (Nugroho and Nurfauzi 2021a; Nugroho et al. 2021), affine transformations (Nugroho and Nurfauzi 2021a; Nugroho et al. 2021), Euclidean geometry (Nugroho and Nurfauzi 2021a), blurring (Nugroho and Nurfauzi 2021a), contrast enhancement (Nugroho and Nurfauzi 2021a; Nugroho et al. 2021), and color transformation (Nugroho and Nurfauzi 2021b).
DL for object detection algorithms has two kinds of approaches, namely one-stage and two-stage detectors. These algorithms are widely utilized to overcome the limitations in diverse medical fields, especially in medical image analyses, including the detection of organs and their abnormalities (Liu et al. 2018; Tang et al. 2018;Lemay 2019), detection and segmentation of thyroid in ultrasound images (Nugroho et al. 2021), detection of pulmonary diseases (Ma et al. 2019; Sirazitdinov et al. 2019; Xiao et al. 2019), detection of cancers (Ezhilarasi and Varalakshmi 2018; van Rijthoven et al. 2018; Liu et al. 2019; Ünver and Ayan 2019), detection and segmentation hemorrhages (Chang et al. 2018), as well as classification and segmentation of microscopy images (Lo et al. 2018; El-Melegy et al. 2019; Kutlu et al. 2020; Nakasi et al. 2021).
Faster R-CNN, a famous DL architecture for object detection using a two-stage detector (Ren et al. 2017), was applied to detect and classify malaria parasites in thin blood smears (Hung et al. 2018). This method was evaluated on a public dataset containing 1300 microscopic thin blood smear images. However, the database only contains one parasite variant (P. Vivax) with four stages (ring, trophozoite, schizont, and gametocyte). The proposed method (Hung et al. 2018) had low results in detection results for trophozoite and ring classes. Moreover, due to the similarity in shape and intensity between malaria parasites and artifacts, this model produced a high rate of FP results for both classes.
Recently, YOLO, the most popular algorithm for object detection has shown good performance in malaria parasite detection. Chibuta and Acar (2020) applied YOLO v3 to detect malaria parasites in two thick blood smear datasets. Their study found that the YOLO-based algorithm had a better performance for malaria detection compared to the previous one (SW + CNN). In another study, Abdurahman et al. (2021) compared YOLO v3, YOLO v4, SSD, and faster R-CNN to detect malaria parasites on thick blood smears. The results showed that both YOLO versions performed better than the other two architectures.
DL for semantic segmentation is a technique to segment objects on an image. Some familiar architectures have been proposed, including FCN (Long et al. 2015), U-Net (Ronneberger et al. 2015), and DeepLab (Chen et al. 2018a). Ronneberger et al. (2015) proposed an architecture called U-Net, which has become famous because it performs well in many medical image applications, such as breast tumor segmentation (Robin et al. 2021) and brain tissue segmentation in MR images (Woo and Lee 2021). Nautre et al. (2020) also applied U-Net to segment malaria parasites on thin blood smear by combining a preprocessing technique for malaria parasite film called GGB (green–green–blue) normalization. They also compared with other color spaces, such as RGB and HSV. The result shows that using GGB normalization is the best combination. They achieved a good performance on the accuracy of 0.995. However, they only inform accuracy, which does not represent oversegmentation and undersegmentation even when the image contains small objects compared to its background.
He et al. (2016) proposed an improved U-Net called Res-UNet, which won first place in the ImageNet competition. The architecture resulted from combining U-Net and ResNet with some modifications was aimed to solve the deep gradient degradation problem. The modification consisted of the following process: (1) designing the convolutional layer, residual unit, and polling layer by adopting the basic concept of ResNet; (2) designing a feature extractor on upsampling and downsampling layers using residual concept motivated by ResNet; (3) applying a linear interpolation technique in the deconvolution step; and (4) adjusting the number of output classes according to user needs.
ResFCN-18 is an improved FCN architecture (Zhu et al. 2020). The large input image required three times downsampling of the feature map and deconvolution layer to get small image patches. Therefore, this architecture modification was based on the feature extractor models on the feature maps. They used three feature extractors. One of them applied ResNet-18 architecture. A recent semantic segmentation model is DeepLabV3, proposed by Chen et al. (2018b). This model used a new feature map extractor named atrous convolution. This convolution model aims to minimize spatial information lost during extracting feature extractor by convoluting and pooling using the traditional manner.
As mentioned earlier, some significant issues to solve in this study are described below.
1.
Some DL models for object detection and semantic segmentation have promised results in many applications. However, the applications in malaria parasite fields are still infrequently explored. Therefore, this study explores some recognized DL architectures in object detection and semantic segmentation on our challenging dataset.
2.
We proposed a new hybrid method by combining optimized threshold and DL-based techniques so that the method can run well for malaria parasite detection and segmentation in thin blood smears images. We also compared our proposed method with the famous architectures in DL for object detection and semantic segmentation.
2. Method and materials
2.1. Data
The PlasmoID dataset (Nugroho et al. 2022) consisted of hundreds of microscopy slides from the Eijkman Institute and Parasitology Laboratory of Universitas Gadjah Mada. Most slides provided by Eijkman Institute were collected from rural areas in Indonesia. These slides contain many artifacts, such as dust caused during staining and storage processes. Figure 1 shows four kinds of the photos. Figure 1a presents a good slide indicated by fairly distributed RBCs, focused objects, and less artifacts. On the other hand, Figs. 1b–1d show our slides containing many artifacts, blurred objects, and clumped RBCs.
Fig. 1.

Figure 1e illustrates the procedure of data collection. The microscopic images were generated by capturing several fields of view containing parasites on a thin blood smear film without overlapping. We used a USB Optilab camera with a 1000× microscope magnification to capture the fields with the output size image of 1600 × 1200 pixels. The dataset consists of 468 fields of view from hundreds of slides with 691 parasites (Nugroho et al. 2022). After collecting the data, a parasitologist from the Department of Parasitology, Universitas Gadjah Mada annotated each parasite's boundary and its classes.
2.2. Proposed method
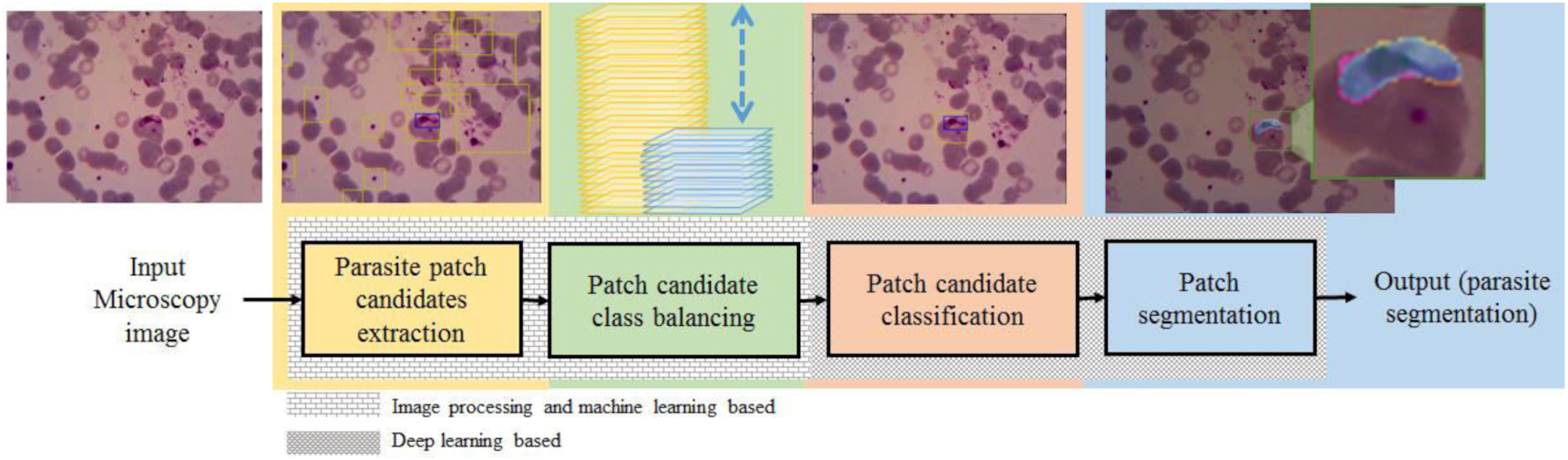
This study aims to solve the issues of detecting malaria parasites in endemic areas in Indonesia's rural areas. Therefore, this study employed a malaria dataset collected from the targeted areas to evaluate our proposed method's performance. We proposed a hybrid malaria detection using image processing approach and DL techniques to run well on our dataset with limited number of images containing many artifacts. As shown in Fig. 2, the proposed method consists of four main steps, extracting parasite patch candidates using image processing techniques, balancing patch candidates using a machine learning technique, classifying patch candidates (parasite detection), and segmenting patch (parasite segmentation) using a DL technique. The different texture backgrounds indicate the different techniques used. The detailed proposed methods are explained as follows.
Fig. 2.
2.2.1. Parasite patch candidate extraction
The objective of this step is to generate malaria parasite patch candidates for object detection and classification to reduce the computation time instead of using sliding windows strategy. There are three primary stages to generating malaria parasite candidate patches shown in Fig. 3, image processing, calculating a global optimum threshold value, and setting patches from labeled segmented areas.
Fig. 3.
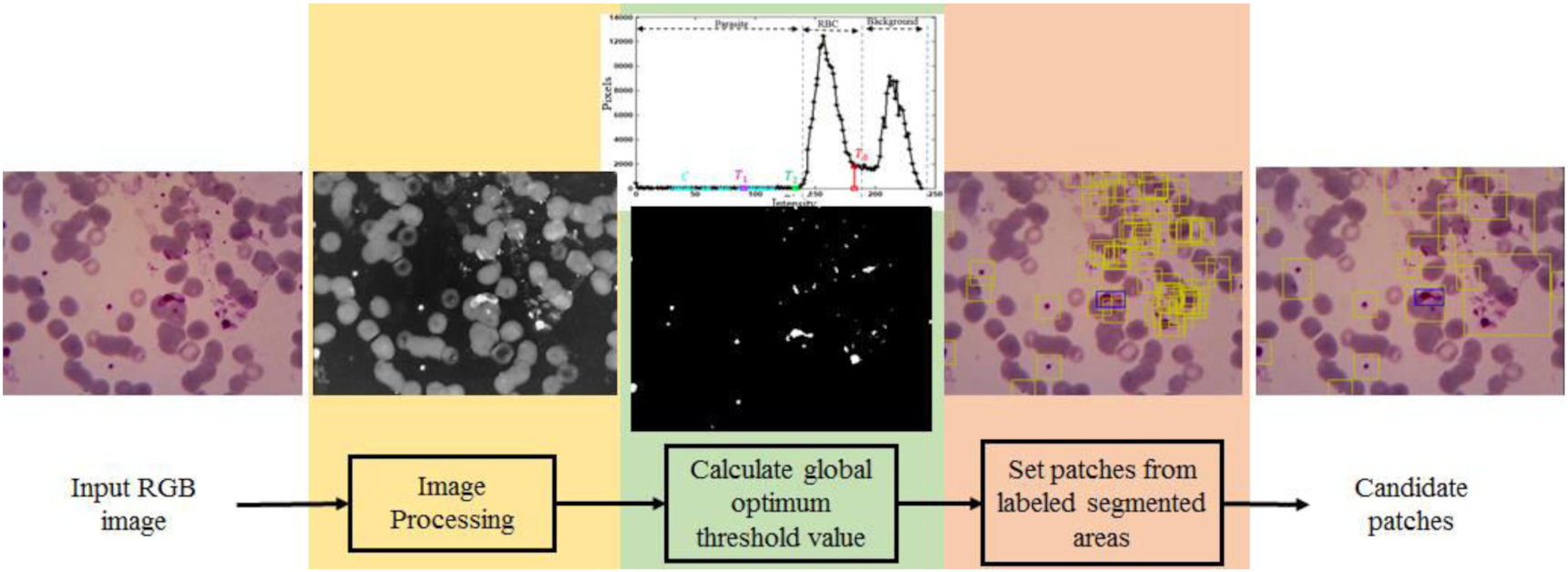
In the image processing stage, the image was prepared to have low noise in four steps, including selecting a green channel, resizing the image resolution, linear contrast stretching, and image complement. By analyzing our data utilizing RGB (red, green, and blue) color space, we chose green channel in our pre-processing step following up the results of previous studies. Studies of Ross et al. (2006) and Rosado et al. (2017) showed that the low-intensity objects, including malaria parasites, appeared clearly with minimum luminance noise. After choosing the green channel, we resized the image into half of the original one, whose resolution was 1600 × 1200. We observed the morphology of malaria parasites on the resized image to ensure no crucial missing information. Resizing step is aimed to reduce the computational time. The linear image stretching technique was applied to enhance the image contrast. Finally, image complement was conducted to highlight the appearance of the parasites.
The second stage was calculating a global threshold value. In this stage, we proposed a new optimized method based on a double Otsu technique to determine an optimum global threshold value capable of adaptively separating the object's intensity and the background's intensity. For example, as shown in Fig. 4, a captured image from a thin blood slide can be classified based on intensity into three classes, low, medium, and high. The low-intensity objects are parasites, WBCs, and some artifacts. On the other hand, medium- and high-intensity classes are commonly only RBCs and a liquid background. Therefore, we need to find three global threshold values to segment the classes in a fast and accurate manner.
Fig. 4.
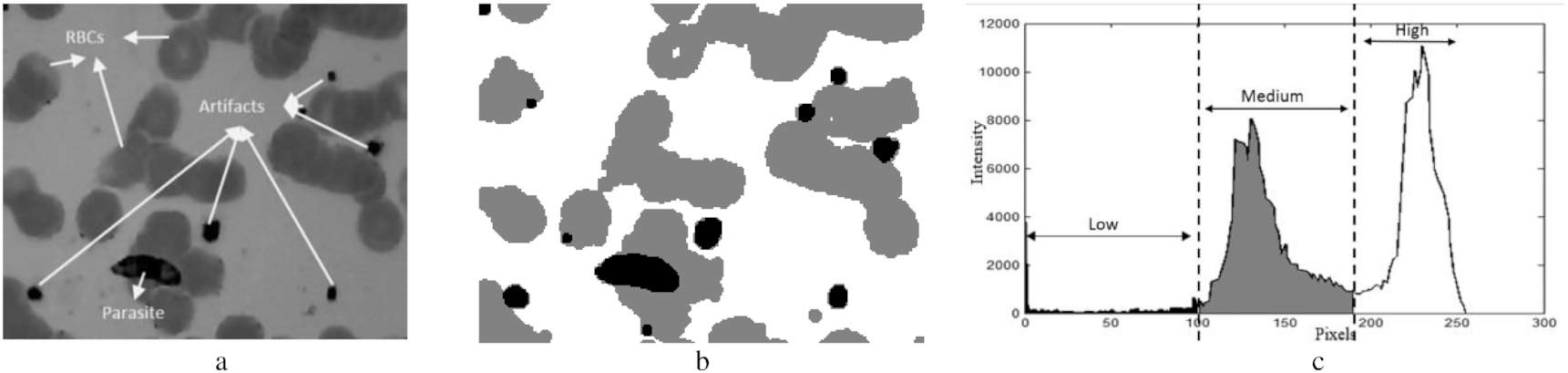
Fast and accurate are two of Otsu's strengths making this method a popular technique for separating objects and backgrounds in an image. The method obtains one or more global threshold values utilized to separate their objects by calculating the variance of intraclass intensity. The modified Otsu method (Memeu et al. 2013) has been applied to segment parasites, RBCs, and background on a microscopy image. However, this method has low accuracy since the image contains a significantly small low- or high-intensity area. Thus, we proposed a technique to find the optimum global threshold values on the modified Otsu method. The steps of the modified Otsu method are shown in Fig. 5 (Memeu et al. 2013). T0 and T1 are global threshold values to threshold RBCs and low-intensity objects, including parasites and artifacts. H1 is a set of the first until T0 of H0.
Fig. 5.

As previously mentioned, regarding the limitation of T1, we proposed a technique to find the optimum global threshold value (T2). The general steps are shown in Fig. 6, and the detailed steps are explained as follows: the first step was to calculate a C graph. The C graph was constructed by summing the mean and standard deviation of each three points of H1. The second step was to calculate a UN and the derivative of the UN. The UN was a binary pattern of C elements thresholded by the average of C. Finally, T2 was defined by the last location of the UN element having one value. UN required an update to avoid missed RBC intensity predictions iteratively using Algorithm 1. The next step was to threshold the input image with the global threshold value (T2) to generate the candidate parasite areas. Finally, each center of the candidate area was calculated to generate the patch-sized 80 × 80 pixels. This number was obtained by observing the RBC size.
Fig. 6.

Algorithm 1.
| j <= the position of nonzeros component of UN’ |
| While st is 0 |
| For i = 1, 2, 3,…, n − 1/do |
| If subtraction of UN’ (j(i)) by UN’ (j(i + 1)) is −2/do |
| If subtraction of j(i + 1) by j(i) is more than 0.2 of z and |
| STD of C on j(i) up to j(i + 1) component is more than STD of C on the first up to j(i + 1)/do |
| • Update UN by carrying the first until the j(i) component of UN |
| • st <= 1 |
| STD = standard deviation |
| n = the number of j component |
| z = the number of H1 component |
2.2.2. Balancing parasite patch candidates
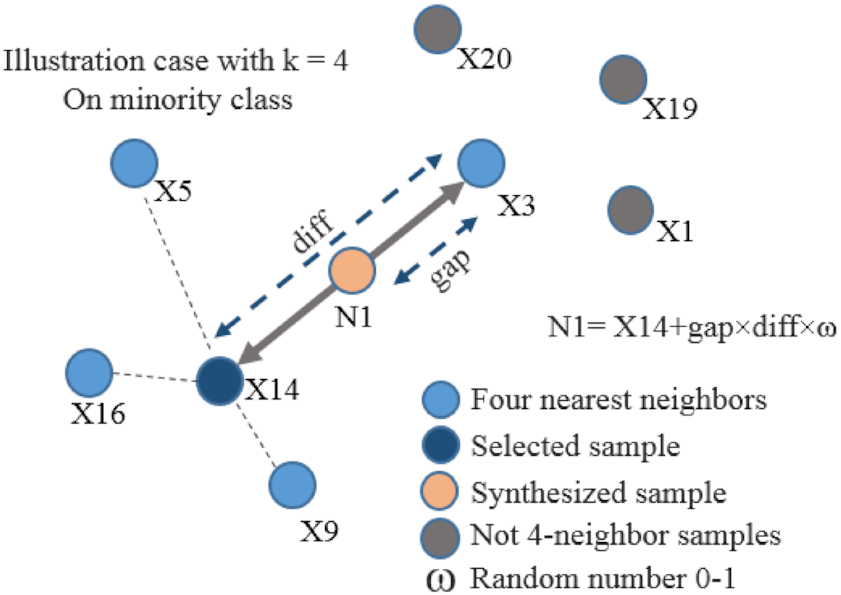
The output of previous step is parasite patch candidates. The candidates were determined only by intensity feature. Consequently, other objects similar in intensity to parasites, such as dust and pellets, were also recognized as parasites. These objects were called as artifacts. An object classification step is needed to eliminate these artifacts. However, because the number of artifacts is far more than parasites, the resulting patch is not balanced between parasites and artifacts. These unbalanced classes would worsen the statically learning models in classification because the trained model was biased towards the majority class (Krawczyk 2016). Various strategies have been conducted to tackle the issues of unbalanced data in the last decade (Kaur et al. 2019). Our study applied one of the earliest and most popular algorithms in oversampling, namely the synthetic minority oversampling technique (SMOTE) (Chawla et al. 2011), to balance our patch classes. SMOTE implemented the K-nearest neighbors’ algorithm to generate a synthetic minority class observation. Then, the observations of the minority class were added continuously until the classes were balanced as illustrated in Fig. 7. There are five steps in the SMOTE process (Chawla et al. 2011), namely,
1.
Identifying the feature vector and its nearest neighbor.
2.
Calculating the difference between the feature vector and a chosen nearest neighbor.
3.
Multiplying the difference with a random number between 0 and 1.
4.
Determining a new point on the line segment by adding the random number to the feature vector.
5.
Repeating the process for identified feature vectors.
Fig. 7.
2.2.3. Patch candidate classification (parasite detection)
After obtaining the balanced parasite patch candidate classes, we needed to classify the patches into parasites and artifacts. The data were divided into two proportions, including training and testing, with a ratio of 8:2. Several light DL models for image classification were observed in this step to find the best model for parasite patch detection. The models were GoogleNet (Szegedy et al. 2015), DenseNet121(Huang et al. 2017), MobileNet V2 (Sandler et al. 2018), MnasNet (Tan et al. 2019), ShuffleNet v2 (Ma et al. 2018), and ResNet50 (He et al. 2016). The best model was applied as artifact patch removal. Some identical pre-processing steps were conducted in every training step in these models. The steps were augmentation and normalization. The detail of these steps is explained as follows:
We constructed a simple augmentation scheme so that the generated images were unique in every epoch but had no substantial difference from the original. We used horizontal and vertical flips and some affine techniques, such as scaling, shearing, and rotating. The flow of the augmentation scheme is presented in Pseudocode 1. After augmenting the image, the color image was normalized by the normalization procedure used in PyTorch n.d.-b.
Pseudocode 1.
| INITIALIZE i = 0 |
| DO FOR all images in the data training |
| INITIALIZE Flip_method = choose randomly one of {horizontal, vertical} |
| INITIALIZE Afine_methods = |
| {Scale: weight Scale ← randomly selected for each axis from 0.9 to 1.1, |
| Translate: weight Scale ← randomly selected for each the axis from −0.01 to 0.01, |
| Shear: weight Scale ← randomly selected from −0.5 to 0.5, |
| Rotate: weight Scale ← randomly selected from −3 to 3} |
| INITIALIZE Lamda = randomly CHOOSE half of an array of 0 to the number of images |
| INPUT the next image |
| Image ←Flip_method (Image) |
| IF i in lamda THEN |
| Image ← Image Afine_methods (Image) |
We trained the models using PyTorch (PyTorch n.d.-a) library. We loaded 32 patches for each epoch for training the model. A standard averaged stochastic gradient descent technique was applied to optimize the training process with learning rate and momentum set to 0.001 and 0.9. The learning rate was reduced by 0.1 when the training processed plateaus. We used a graphics processing unit built-in GeForce GTX 1050Ti manufactured by NVIDIA to speed up the training process, which required high computations when training DL models. The trained model could smoothly run to predict an image on low-resource client devices. Generally, the model was trained once or more depending on the model's performance in the real world. The output of these processes was patch candidates. Our detection approach was compared to two famous DL models for object detection, namely faster R-CNN (Ren et al. 2017) and Yolo v5s (van Rijthoven et al. 2018).
2.2.4. Patch segmentation (parasite segmentation)
After we got a patch classified as a parasite patch or region of interest (ROI) of the parasite, we segmented it using DL techniques for semantic segmentation. In this section, we employed five DL models to show the difference between applying ROI in the segmenting process and not applying it. The models were UNet (Ronneberger et al. 2015), Res-UNet (He et al. 2016), ResFCN (Zhu et al. 2020), DeepLabV3 (Chen et al. 2018a), and DeepLabV3+ (Chen et al. 2018a). The training processes were similar to the patch candidate classification process, such as resizing, augmenting, normalizing, and transferring learning. The output of this process was segmented parasites.
2.3. Performance evaluation
Three performance evaluations that can represent the performance of our goal are precision, recall, and F1-score. Precision defines how the system or model successfully detects positive class predictions that actually belong to the positive class. Recall defines how the system successfully detects the number of positive class predictions made out of all positive examples in all patch candidates. Meanwhile, F1-score delivers a performance score that balances both the concern of precision and recall. In addition, this study also considers the performance of computational time, representing high computations to detect the parasites. Precision, recall and F1-score are mathematically formulated in the eqs. 1, 2, and 3 (Chibuta and Acar 2020). The output is either a parasite (+ve) or non-parasite (−ve), with only two classes of detection and segmentation class.
True positive (TP): prediction is +ve, and X is a parasite; we want that
True negative (TN): prediction is −ve, and X is a non-parasite; we want that too
False positive (FP): prediction is +ve, and X is non-parasite; false alarm, bad
False negative (FN): prediction is −ve, and X a parasite; the worst
(1)
(2)
(3)
3. Results and discussion
This study proposes a hybrid scheme to detect and segment malaria parasites by combining double Otsu optimization, machine learning, and DL techniques. A method of optimizing the double Otsu to find the global threshold value for parasites is introduced in this work to generate patch candidates. This strategy aims to overcome the limitation of our dataset, including a small dataset containing many artifacts. This section presents results of each step of our scheme. The proposed scheme consists of four steps, which are extraction, balancing, classification of parasite patch candidates (parasite detection), and parasite segmentation.
In extracting parasite patch candidates, we proposed an optimization of the previous global threshold value to improve the accuracy. The global threshold value was originally proposed by Memeu et al. (2013) to segment objects with a similar intensity to the parasite. However, Memeu et al. (2013) applied the first Otsu to obtain the first global threshold value (T0) for determination of RBC's intensity. Moreover, the second Otsu was applied to obtain the second global threshold value (T1) for determination of the parasite's intensity. Consequently, these rules are unsuitable for our dataset, with a wide RBC intensity range. As a result, implementing Memeu et al. (2013) on our dataset yielded a high FP of 43 678, as shown in Table 1. An example of segmented objects obtained by T1 is shown in Fig. 8a indicated by green contours. However, the segmented objects are not only the parasites but also some clumped RBCs. It is because the second threshold value (T1) is still within the intensity range of the RBC, as shown in Fig. 8b. As a result, the generated patches are massive, as shown in Fig. 8c.
Fig. 8.
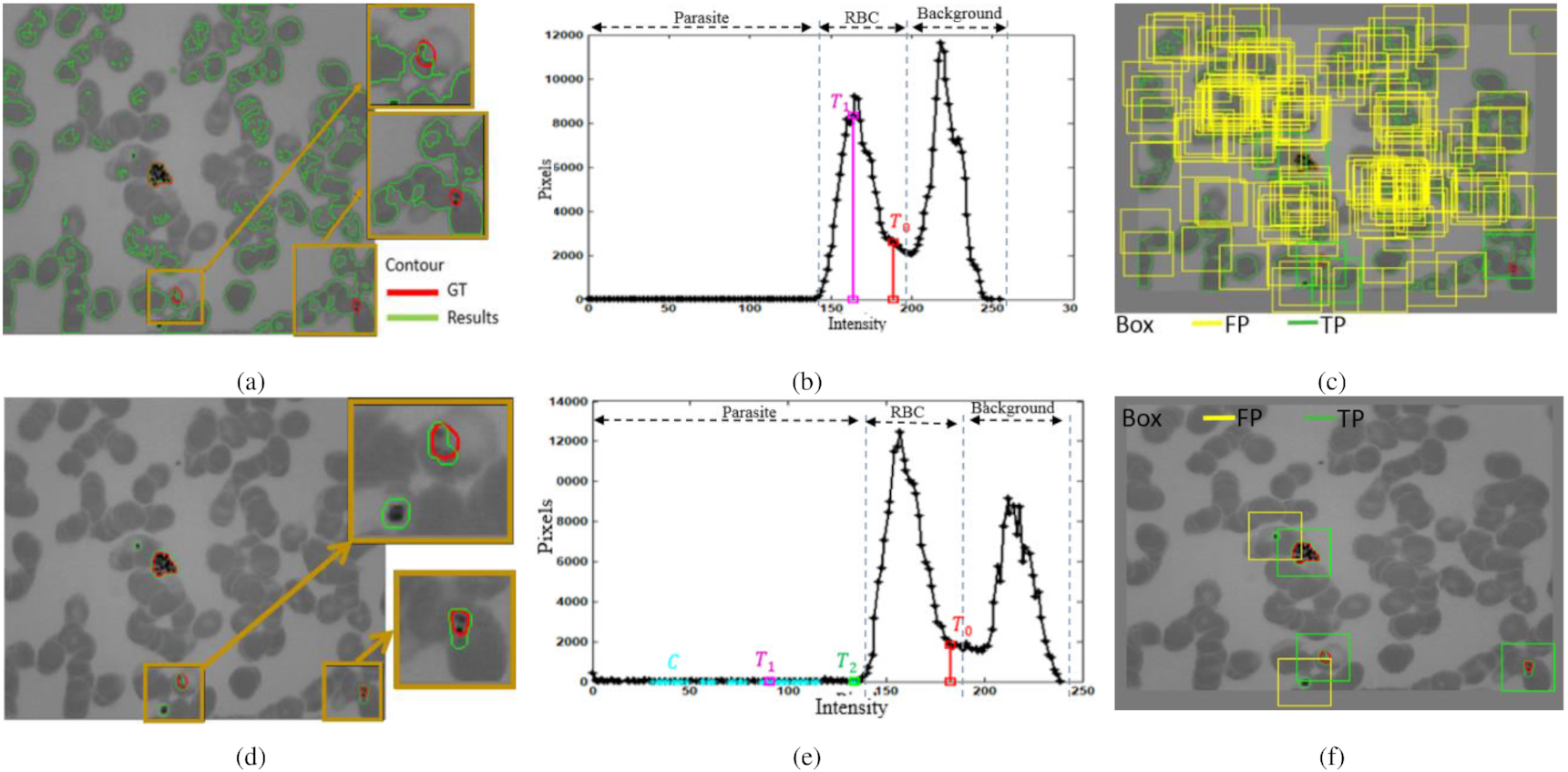
Table 1.
| Methods | TP | FN | FP | Time (s) | Sensitivity (%) |
|---|---|---|---|---|---|
| Nugroho et al. (2017) | 569 | 122 | 1340 | 0.489 | 82.3 |
| Memeu et al. (2013) | 677 | 14 | 43 678 | 0.039 | 98 |
| Proposed method | 688 | 3 | 3555 | 0.073 | 99.6 |
After using our optimized method, the sensitivity can be increased from 98% to 99.6%. In order words, there were only three parasites missed out of 691 parasites. Moreover, our proposed method successfully suppressed the number of FPs from 43 678 to 3555 but is still higher than by Nugroho et al. (2017). The objects can be segmented more accurately (Fig. 8d) because the optimized second threshold value (T2) lies at the edge of parasite's and RBC's intensities, as shown in Fig. 8e. As a result, the generated patches successfully show more accurate parasites and significantly reduce FP parasite patches.
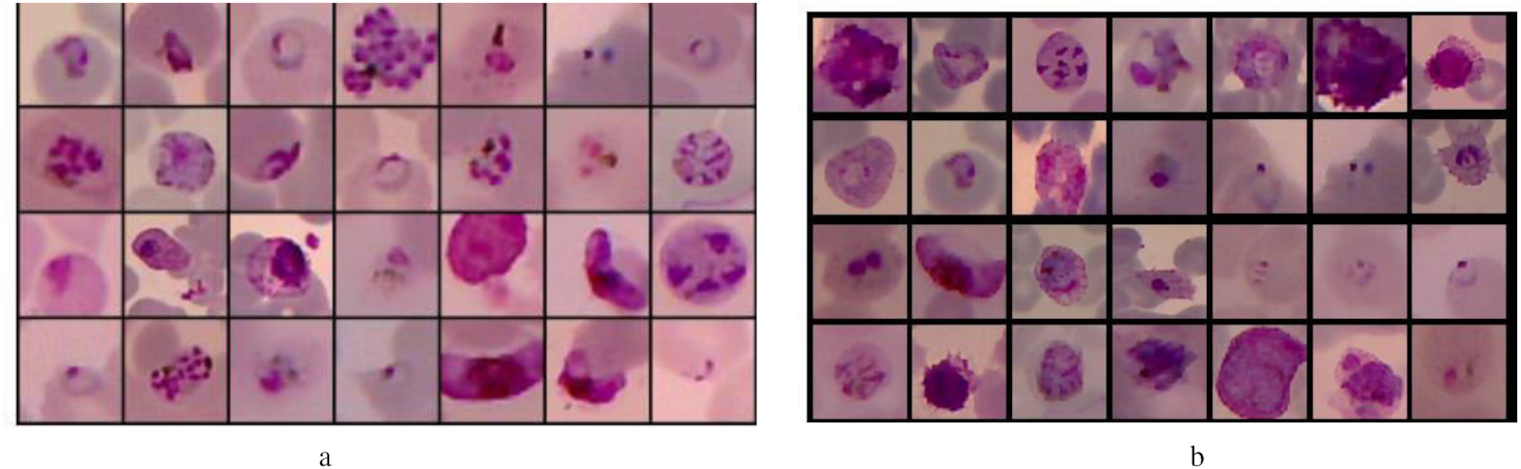
The generated patches as the output of this process still contained many FPs. Therefore, we needed to eliminate it using a DL technique for image classification. Furthermore, because the generated patches contained too many FP compared to the parasites, we should balance them to avoid the trained model being biased towards the majority class. Here, we used SMOTE (Chawla et al. 2011) to balance both classes. The image in the minority class, the parasite, was augmented to the number of patches in the FP class, 3555. The samples of the patch augmentation results are shown in Fig. 9.
Fig. 9.
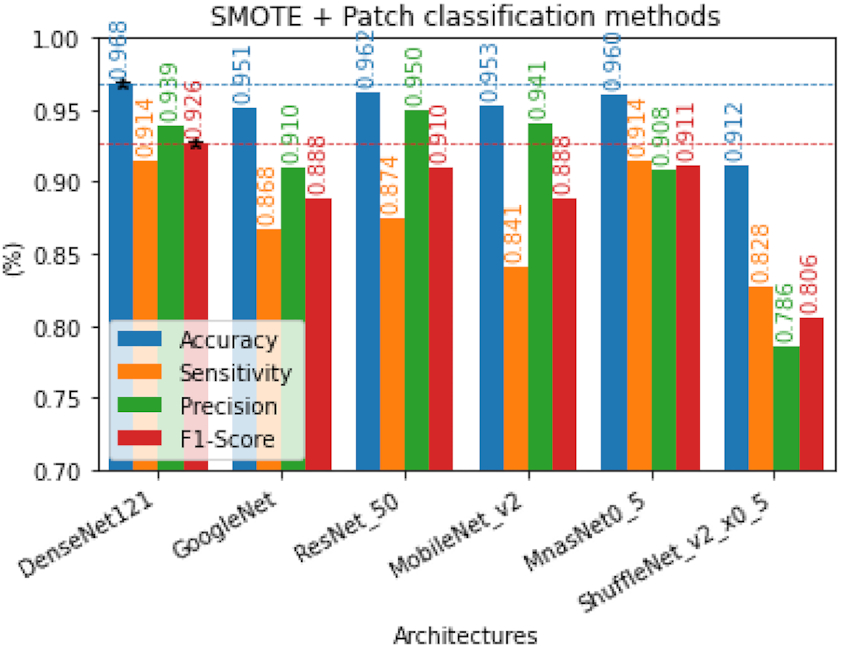
After both classes were balanced, we classified them using DL for image classification techniques. This study explored six models of DL for image classification, namely, GoogleNet (Szegedy et al. 2015), DenseNet121(Huang et al. 2017), MobileNet V2 (Sandler et al. 2018), MnasNet (Tan et al. 2019), ShuffleNet v2 (Ma et al. 2018), and ResNet50 (He et al. 2016). This step aimed to find the best DL model for FP reduction or the end step of parasite detection in thin blood smear film. These comparative study results are shown in Fig. 10. F1-score is the most considered performance in selecting the models because it combines specificity and precision by taking their harmonic means. Thus, we assume that DenseNet has the best performance in recognizing parasites and artifacts.
Fig. 10.
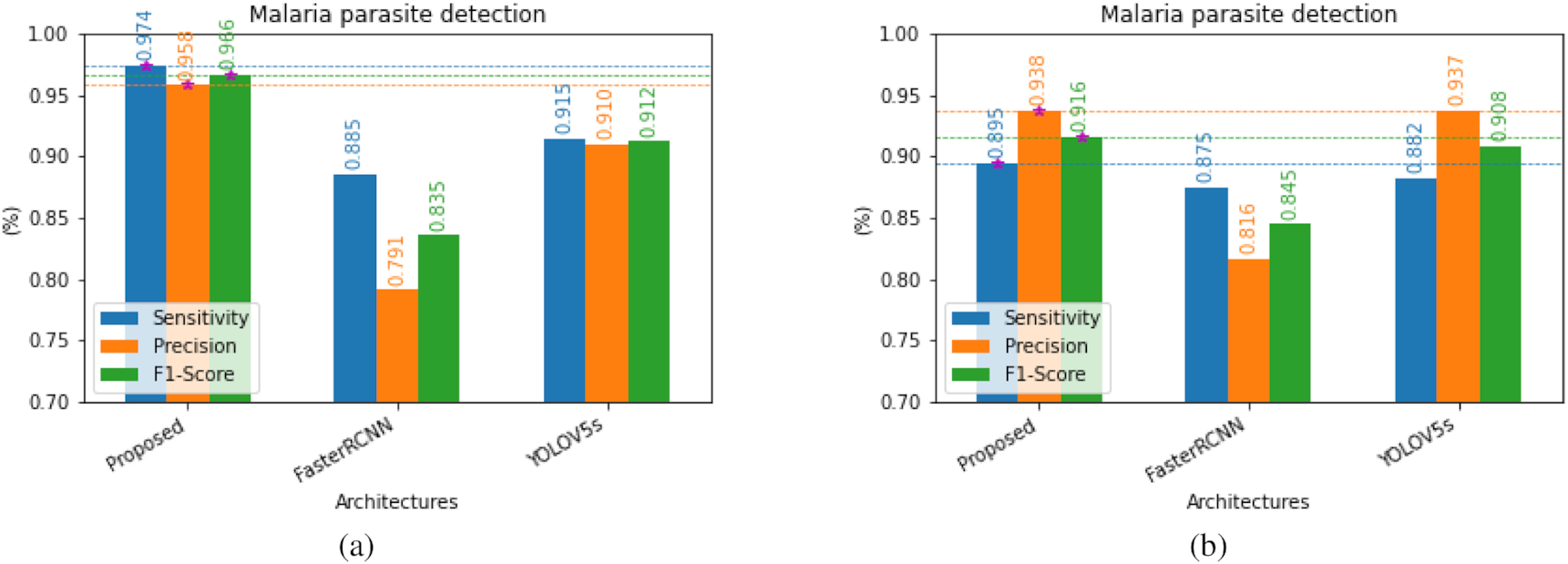
The proposed hybrid scheme combines double Otsu optimization, machine learning, and DL techniques to improve the performances of parasite detection and segmentation. This scheme was also compared with two familiar models in DL for object detection, namely, Yolo v5 light/small version (van Rijthoven et al. 2018) and Faster R-CNN with FPN ResNet50 (Ren et al. 2017). We limited the performance evaluation to three, i.e., sensitivity or recall, precision, and F1-score. The performance evaluation results of the data training and testing are shown in Figs. 11a and 11b. Referring to Fig. 11b, the proposed method has significantly better results than faster R-CNN (Ren et al. 2017) and relatively better results than Yolo v5s (van Rijthoven et al. 2018) on the data testing. However, the proposed method is significantly better than both methods in the data training. These results indicate that the proposed method performs better in detecting malaria parasites in our dataset with a limited number of images containing many artifacts. It is difficult to distinguish between parasites and artifacts when observing a small area due to the fact that certain types of tiny parasites possess similar characteristics to artifacts, which is the key reason behind this challenge. Hence, the proposed method enlarges the area of patches up to RBC size when a patch is more petite than RBC size.
Fig. 11.
The next step was parasite segmentation. This step aimed to get the morphology of parasites in more detail to support the advanced malaria parasite studies. After we got the parasite location, we segmented it using DL models for semantic segmentation. We explored five contemporary architectures to segment a parasite on the patch or ROI, and then we compared it without using ROI. The architectures were UNet (Ronneberger et al. 2015), Res-UNet (He et al. 2016), ResFCN (Zhu et al. 2020), DeepLabV3 (Chen et al. 2018a), and DeepLabV3+ (Chen et al. 2018a). The comparison results are depicted in Fig. 12. The proposed scheme performs better in all parameter comparisons than the previous. It implies that finding ROI or object detection in the beginning step of parasite segmentation improves the performances of all algorithms compared without finding ROI. Analyzing an image pixel by pixel is more difficult than analyzing it patch by patch since certain parasites show similar characteristics as artifacts when observed pixel by pixel. Our hypotheses are proven in the comparison results.
Fig. 12.

4. Conclusion
This study develops a hybrid scheme to detect and segment malaria parasites by combining double Otsu optimization method and DL techniques. An optimized double Otsu method for generating malaria parasite patch candidates was proposed. The parasites were then recognized and segmented using DL approaches. Having tested on the PlasmoID dataset containing 691 malaria parasites, the proposed scheme successfully achieved an F1-score of 0.91 for parasite detection. The proposed scheme also outperformed original semantic segmentation methods in terms of sensitivity, specificity, and F1-score for parasite segmentation. This finding indicates that the proposed scheme has a potential to be implemented in the malaria computer-aided detection system. In the next work, an advance of optimized local threshold method can be further developed for the microscopic images consisting severe luminance noises.
Acknowledgment
The authors express their gratitude to the Department of Electrical and Information Engineering at Universitas Gadjah Mada for providing valuable research facilities and to the Research Grant of “World Class Research” from The Deputy of Research and Development at the National Research and Innovation Agency Republic of Indonesia for their generous funding support.
References
Abdurahman F., Fante K.A., Aliy M. 2021. Malaria parasite detection in thick blood smear microscopic images using modified YOLOV3 and YOLOV4 models. BMC Bioinformatics, 22(1): 112.
Anon. 2018. WHO | World Malaria Report 2018. WHO.
Bates I., Bekoe V., Asamoa-Adu A. 2004. Improving the accuracy of malaria-related laboratory tests in Ghana. Malaria Journal, 3(1): 38.
Chang P.D., Kuoy E., Grinband J., Weinberg B.D., Thompson M., Homo R., et al. 2018. Hybrid 3D/2D convolutional neural network for hemorrhage evaluation on head CT. American Journal of Neuroradiology, 39(9): 1609–1616.
Chawla N.V., Bowyer K.W., Hall L.O., Kegelmeyer W.P. 2011. SMOTE: synthetic minority over-sampling technique. Journal of Artificial Intelligence Research, 16: 321–357.
Chen L.-C., Papandreou G., Kokkinos I., Murphy K., Yuille A.L. 2018a. DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Transactions on Pattern Analysis and Machine Intelligence, 40(4): 834.
Chen L.-C., Zhu Y., Papandreou G., Schroff F., Adam H. 2018b. Encoder–decoder with atrous separable convolution for semantic image segmentation. In Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics) 11211. pp. 833–851.
Chibuta S., Acar A.C. 2020. Real-time malaria parasite screening in thick blood smears for low-resource setting. Journal of Digital Imaging, 33(3): 763–775.
Dave I.R., Upla K.P. 2017. Computer aided diagnosis of malaria disease for thin and thick blood smear microscopic images. In 2017 4th International Conference on Signal Processing and Integrated Networks (SPIN). pp. 561–565.
El-Melegy M., Mohamed D., ElMelegy T. 2019. Automatic detection of tuberculosis bacilli from microscopic sputum smear images using faster R-CNN, transfer learning and augmentation. In Pattern recognition and image analysis: 9th Iberian Conference, IbPRIA 2019, Madrid, Spain, 1–4 July 2019, Proceedings, Part I. Springer-Verlag, Berlin, Germany. pp. 270–278.
Ezhilarasi R., Varalakshmi P. 2018. Tumor detection in the brain using faster R-CNN. In 2018 2nd International Conference on I-SMAC (IoT in Social, Mobile, Analytics and Cloud). I-SMAC (IoT in Social, Mobile, Analytics and Cloud). pp. 388–392.
Frean J. 2010. Microscopic determination of malaria parasite load: role of image analysis. In Microscopy: Science, Technology, Applications and Education. pp. 862–866.
He K., Zhang X., Ren S., Sun J. 2016. Deep residual learning for image recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. Vol. 2016-December. IEEE Computer Society. pp. 770–778.
Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. 2017. Densely connected convolutional networks. In Proceedings - 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017. Vol. 2017-January. Institute of Electrical and Electronics Engineers Inc.pp. 2261–2269.
Hung J., Ravel D., Lopes S.C.P., Rangel G., Nery O.A., Malleret B. et al., 2018. Applying faster R-CNN for object detection on malaria images. In IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops 2017-January.
Kaur H., Pannu H.S., Malhi A.K. 2019. A systematic review on imbalanced data challenges in machine learning. ACM Computing Surveys, 52(4): 1.
Krawczyk B. 2016. Learning from imbalanced data: open challenges and future directions. Progress in Artificial Intelligence, 5(4): 221–232.
Kutlu H., Avci E., Özyurt F. 2020. White blood cells detection and classification based on regional convolutional neural networks. Medical Hypotheses 135: 109472.
Lemay A. 2019. Kidney recognition in CT using YOLOv3. Available from https://doi.org/10.48550/arXiv.1910.01268.
Liu M., Jiang J., Wang Z. 2019. Colonic polyp detection in endoscopic videos with single shot detection based deep convolutional neural network. IEEE Access, 7: 75058–75066.
Liu W., Cheng L., Meng D. 2018. Brain slices microscopic detection using simplified SSD with cycle-GAN data augmentation. In BT - neural information processing. Edited by L. Cheng, A.C.S. Leung, S. Ozawa. Springer International Publishing, Cham, Switzerland. pp. 454–463.
Lo Y.-C., Juang C.-F., Chung I.F, Guo S.-N., Huang M.-L., Wen M.-C., et al. 2018. Glomerulus detection on light microscopic images of renal pathology with the faster R-CNN. In BT - neural information processing. Edited by L. Cheng, A.C.S. Leung, S. Ozawa. Springer International Publishing, Cham, Switzerland. pp. 369–377.
Loddo A., Di Ruberto C., Kocher M. 2018. Recent advances of malaria parasites detection systems based on mathematical morphology. Sensors, 18(2): 513.
Long J., Shelhamer E., Darrell T. 2015. Fully convolutional networks for semantic segmentation. In 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE Computer Society, Los Alamitos, CA. pp. 3431–3440.
Ma J., Li X., Li H., Menze B.H., Liang S., Zhang R., Zheng W.-S. 2019. Group-attention single-shot detector (GA-SSD): finding pulmonary nodules in large-scale CT images. In Proceedings of the 2nd International Conference on Medical Imaging with Deep Learning. Vol. 102. Edited by M.J. Cardoso, A. Feragen, B. Glocker, E. Konukoglu, I. Oguz, G. Unal, T. Vercauteren. PMLR. pp. 358–369.
Ma N., Zhang X., Zheng H.-T., Sun J. 2018. ShuffleNet V2: practical guidelines for efficient CNN architecture design. In Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics) 11218. pp. 122–138.
Maqsood A., Farid M.S., Khan M.H., Grzegorzek M. 2021. Deep malaria parasite detection in thin blood smear microscopic images. Applied Sciences, 11(5): 2284.
Maysanjaya I.Md.D., Nugroho H.A., Setiawan N.A., Murhandarwati E.E.H. 2016. Segmentation of Plasmodium vivax phase on digital microscopic images of thin blood films using colour channel combination and Otsu method. In AIP Conference Proceedings. Vol. 1755. AIP Publishing LLC. p. 160002.
Memeu D.M., Kaduki K.A., Mjomba A.C.K., Muriuki N.S., Gitonga L. 2013. Detection of plasmodium parasites from images of thin blood smears. Open Journal of Clinical Diagnostics, 03(4): 183–194.
Mitiku K., Mengistu G., Gelaw B. 2003. The reliability of blood film examination for malaria at the peripheral health unit. Ethiopian Journal of Health Development, 17(3): 197–204.
Moody A. 2002. Rapid diagnostic tests for malaria parasites. Clinical Microbiology Reviews, 15(1): 66–78.
Moon S., Lee S., Kim H., Freitas-Junior L.H., Kang M., Ayong L., Hansen M.A.E. 2013. An image analysis algorithm for malaria parasite stage classification and viability quantification. PLoS ONE, 8(4): e61812.
Nakasi R., Mwebaze E., Zawedde A. 2021. Mobile-aware deep learning algorithms for malaria parasites and white blood cells localization in thick blood smears. Algorithms, 14(1): 17.
Nautre A., Nugroho H.A., Frannita E.L., Nurfauzi R. 2020. Detection of malaria parasites in thin red blood smear using a segmentation approach with U-Net. in 2020 3rd International Conference on Biomedical Engineering (IBIOMED). pp. 55–59.
Nugroho H.A., Nurfauzi R. 2021a. Convolutional neural network for classifying retinal diseases from OCT2017 dataset. In 2021 4th International Conference on Information and Communications Technology (ICOIACT). pp. 295–298.
Nugroho H.A., Nurfauzi R. 2021b. GGB color normalization and faster-RCNN techniques for malaria parasite detection. In 2021 IEEE International Biomedical Instrumentation and Technology Conference (IBITeC). pp. 109–113.
Nugroho H.A., Saputra W.A., Permanasari A.E., Elsa Herdiana Murhandarwati E. 2017. Automated determination of plasmodium region of interest on thin blood smear images. In 2017 International Seminar on Intelligent Technology and Its Applications (ISITIA). IEEE. pp. 352–355.
Nugroho H.A., Marsiano A.F.D., Xaphakdy K., Sihakhom P., Frannita E.L., Nurfauzi R., Murhandarwati E.E.H. 2019a. Multithresholding approach for segmenting plasmodium parasites. In 2019 11th International Conference on Information Technology and Electrical Engineering, ICITEE 2019
Nugroho H.A., Nurfauzi R., Frannita E.L. 2019b. Plasmodium candidate detection on thin blood smear images with luminance noise reduction. In Proceeding - 2019 5th International Conference on Science in Information Technology: Embracing Industry 4.0: Towards Innovation in Cyber Physical System, ICSITech 2019.
Nugroho H.A., Frannita E.L., Nurfauzi R. 2021. An automated detection and segmentation of thyroid nodules using Res-UNet. In International Conference on Electrical Engineering, Computer Science and Informatics (EECSI) 2021-October. pp. 181–185.
Nugroho H.A., Nurfauzi R., Murhandarwat E.E.H., Purwono P. 2022. PlasmoID: a dataset for Indonesian malaria parasite detection and segmentation in thin Bbood smear. Available from https://arxiv.org/abs/2211.15105v1.
Poostchi M., Ersoy I., McMenamin K., Gordon E., Palaniappan N., Pierce S., et al. 2018. Malaria parasite detection and cell counting for Human and mouse using thin blood smear microscopy. Journal of Medical Imaging, 5(04):1.
PyTorch. n.d.-a. Available from https://pytorch.org/ [accessed 8 May 2022].
PyTorch. n.d.-b. Pytorch documentation. Available from https://pytorch.org/vision/stable/models.html [accessed 8 May 2021].
Quinn J.A., Andama A., Munabi I., Kiwanuka F.N. 2014. Automated blood smear analysis for mobile malaria diagnosis. In Mobile point-of-care monitors and diagnostic device design. Vol. 31. pp. 115–132.
Ren S., He K., Girshick R., Sun J. 2017. Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Transactions on Pattern Analysis and Machine Intelligence, 39(6): 1137–1149.
Robin M., John J., Ravikumar A. 2021. Breast tumor segmentation using U-NET. In 2021 5th International Conference on Computing Methodologies and Communication (ICCMC). pp. 1164–1167.
Ronneberger O., Fischer P., Brox T. 2015. U-Net: convolutional networks for biomedical image segmentation. In BT - medical image computing and computer-assisted intervention – MICCAI 2015. Edited by N. Navab, J. Hornegger, W. M. Wells, A. F. Frangi. Springer International Publishing, Cham, Switzerland. pp. 234–241.
Rosado L., Correia Da Costa J.M., Elias D., Cardoso J.S. 2017. Mobile-based analysis of malaria-infected thin blood smears: automated species and life cycle stage determination. Sensors, 17(10): 2167.
Ross N.E., Pritchard C.J., Rubin D.M., Dusé A.G. 2006. Automated image processing method for the diagnosis and classification of malaria on thin blood smears. Medical and Biological Engineering and Computing, 44(5): 427–436.
Sandler M., Howard A., Zhu M., Zhmoginov A., Chen L.C. 2018. MobileNetV2: inverted residuals and linear bottlenecks. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. IEEE Computer Society. pp. 4510–4520.
Sirazitdinov I., Kholiavchenko M., Mustafaev T., Yixuan Y., Kuleev R., Ibragimov B. 2019. Deep neural network ensemble for pneumonia localization from a large-scale chest x-ray database. Computers & Electrical Engineering, 78: 388–399.
Sultani W., Nawaz W., Javed S., Danish M., Saadia A., Ali M. 2022. Towards low-cost and efficient malaria detection. In 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). IEEE Computer Society, Los Alamitos, CA. pp. 20655–20664.
Szegedy C., Liu W., Jia Y., Sermanet P., Reed S., Anguelov D., et al. 2015. Going deeper with convolutions. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition 7–12 June 2015. IEEE Computer Society. pp. 1–9.
Tan M., Chen Bo, Pang R., Vasudevan V., Sandler M., Howard A., Le Q.V. 2019. Mnasnet: platform-aware neural architecture search for mobile. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition 2019 June. IEEE Computer Society. pp. 2815–2823.
Tang W., Zou D., Yang Su, Shi J. 2018. DSL: automatic liver segmentation with faster R-CNN and DeepLab. In BT - Artificial neural networks and machine learning – ICANN 2018. Edited by V. Kůrková, Y. Manolopoulos, B. Hammer, L. Iliadis, I. Maglogiannis. Springer International Publishing, Cham, Switzerland. pp. 137–147.
Tek F.B, Dempster A.G., Kale I. 2009. Computer vision for microscopy diagnosis of malaria. Malaria Journal, 8(1): 153.
Thimasarn K., Miller R.S, Maneechai N., Sattabongkot J., Soyseng V., Rachaphaew N., et al. 2002. Comparison of field and expert laboratory microscopy for active surveillance for asymptomatic Plasmodium falciparum and Plasmodium vivax in western Thailand. American Society of Tropical Medicine and Hygiene, 67(2): 141–144.
Ünver H.M., Ayan E. 2019. Skin lesion segmentation in dermoscopic images with combination of YOLO and GrabCut algorithm. Diagnostics, 9(3): 72.
van Rijthoven M., Swiderska-Chadaj Z., Seeliger K., van der Laak J., Ciompi F. 2018. “You Only Look on Lymphocytes Once”.
Woo B., Lee M. 2021. Comparison of tissue segmentation performance between 2D U-Net and 3D U-Net on brain MR images. In 2021 International Conference on Electronics, Information, and Communication (ICEIC). pp. 1–4.
Xiao J., Zhang Y., Bian K., Zhou G., Yan W. 2019. DenXFPN: pulmonary pathologies detection based on dense feature pyramid networks. In ICASSP 2019 - 2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP). pp.1234–1238.
Zhu Y., Tang C., Liu H., Huang P. 2020. End-face localization and segmentation of steel bar based on convolution neural network. IEEE Access, 8: 74679–74690.
Information & Authors
Information
Published In

FACETS
Volume 8 • 2023
Pages: 1 - 12
Editor: Ivana Pajic-Lijakovic
History
Received: 20 September 2022
Accepted: 1 March 2023
Version of record online: 17 July 2023
Copyright
© 2023 The Author(s). This work is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Data Availability Statement
All data used in this study were taken from the referred source.
Key Words
Sections
Subjects
Plain Language Summary
Proposed CAD system using image processing and deep learning techniques for detecting malaria
Authors
Author Contributions
Conceptualization: HAN, RN
Data curation: RN
Formal analysis: HAN
Funding acquisition: HAN
Investigation: RN
Methodology: HAN, RN
Project administration: HAN, RN
Resources: HAN, RN
Software: RN
Supervision: HAN
Validation: HAN
Writing – original draft: RN
Writing – review & editing: HAN, RN
Competing Interests
The authors declare that there is no conflict of interest regarding the publication of this paper.
Metrics & Citations
Metrics
Other Metrics
Citations
Cite As
Hanung Adi Nugroho and Rizki Nurfauzi. 2023. A combination of optimized threshold and deep learning-based approach to improve malaria detection and segmentation on PlasmoID dataset. FACETS.
8: 1-12.
https://doi.org/10.1139/facets-2022-0206
Export Citations
If you have the appropriate software installed, you can download article citation data to the citation manager of your choice. Simply select your manager software from the list below and click Download.
Cited by
1. Development of Low Cost, Automated Digital Microscopes Allowing Rapid Whole Slide Imaging for Detecting Malaria
